Taxonomy Assignment
Overview
Teaching: 15 min
Exercises: 30 minQuestions
What are the main strategies for assigning taxonomy to OTUs?
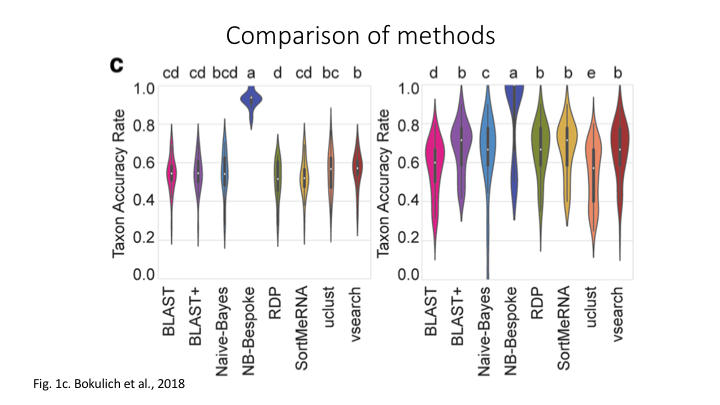
What is the difference between Sintax and Naive Bayes?
Objectives
Assign taxonomy using Sintax
Use
lessto explore a long help outputWrite a script to assign taxonomy
Taxonomy Assignment: Different Approaches
There are four basic strategies to taxonomy classification (and endless variations of each of these): sequence similarity (SS), sequence composition (SC), phylogenetic (Ph), and probabilistic (Pr) (Hleap et al., 2021).
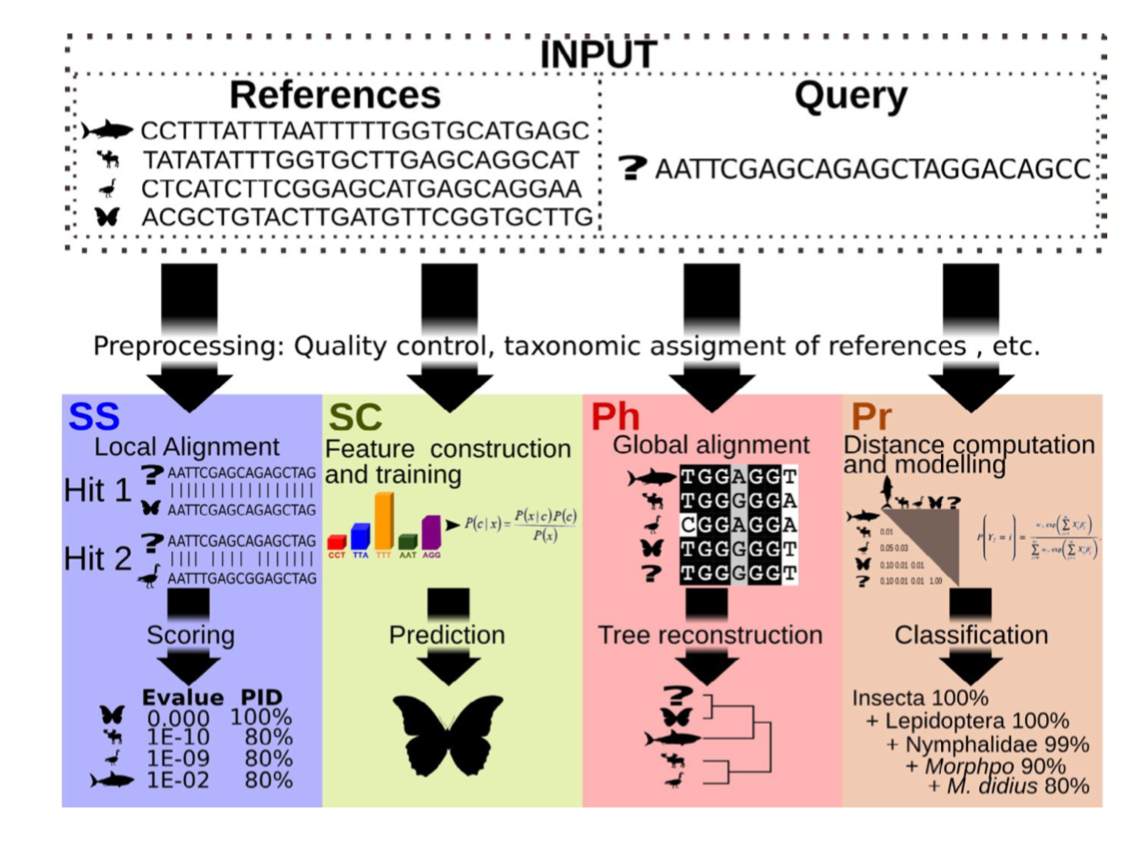
Of the four, the first two are those most often used for taxonomy assignment. For further comparison of these refer to this paper.
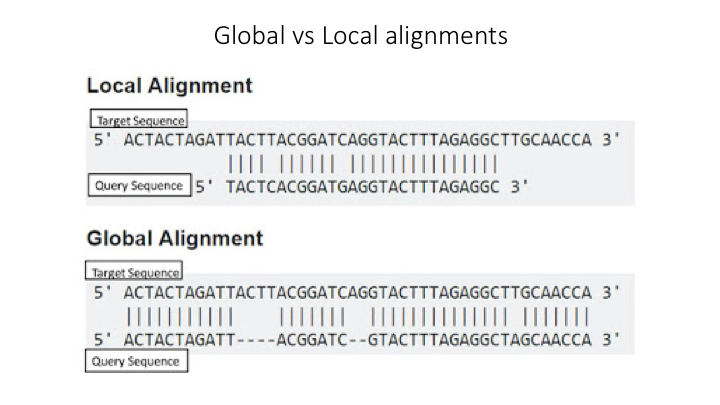
Aligment-Based Methods (SS)
All sequence similarity methods use global or local alignments to directly search a reference database for partial or complete matches to query sequences. BLAST is one of the most common methods of searching DNA sequences. BLAST is an acronym for Basic Local Alignment Search Tool. It is called this because it looks for any short, matching sequence, or local alignments, with the reference. This is contrasted with a global alignment, which tries to find the best match across the entirety of the sequence.
Using Machine Learning to Assign Taxonomy
Sequence composition methods utilise machine learning algorithms to extract compositional features (e.g., nucleotide frequency patterns) before building a model that links these profiles to specific taxonomic groups.
Use the program Sintax to classify our OTUs
The program Sintax is similar to the Naive Bayes classifier used in Qiime and other programs, in that small 8 bp kmers are used to detect patterns in the reference database, but instead of frequency patterns, k-mer similarity is used to identify the top taxonomy, so there is no need for training (Here is a description of the Sintax algorithm). Because of this, it is simplified and can run much faster. Nevertheless, we have found it as accurate as Naive Bayes for the fish dataset we are using in this workshop.
As with the previous steps, we will be using a script to run the classification.
nano taxo
#!/bin/bash
source moduleload
cd ../final/
vsearch --sintax otus.fasta --db ../refdb/fish_16S_sintax.fasta --sintax_cutoff 0.8 --tabbedout taxonomy_otus.txt
chmod +x taxo
./taxo
vsearch v2.17.1_linux_x86_64, 503.8GB RAM, 72 cores
https://github.com/torognes/vsearch
Reading file ../refdb/fish_16S_sintax.fasta 100%
WARNING: 1 invalid characters stripped from FASTA file: ?(1)
REMINDER: vsearch does not support amino acid sequences
12995009 nt in 75207 seqs, min 101, max 486, avg 173
Counting k-mers 100%
Creating k-mer index 100%
Classifying sequences 100%
Classified 22 of 26 sequences (84.62%)
Taxonomy assignment through SINTAX was able to classify 22 out of 26 OTUs. Now that we have the taxonomy assignment file taxonomy_otus.txt and a frequency table otutable.txt, we have all files needed for the statistical analysis.
Study Questions
Name an OTU that would have better resolution if the cutoff were set to default
Name an OTU that would have worse resolution if the cutoff were set higher
Key Points
The main taxonomy assignment strategies are global alignments, local alignments, and machine learning approaches
The main difference is that Naive Bayes requires the reference data to be trained, whereas Sintax looks for only kmer-based matches