Introduction to the bioinformatic analysis of eDNA metabarcoding data
Overview
Teaching: 15 min
Exercises: 0 minQuestions
What are the main components of metabarcoding pipelines?
Objectives
Review typical metabarcoding workflows
eDNA Workshop
Welcome to the environmental DNA (eDNA) workshop session of the Otago Bioinformatic Spring School 2024!
The primary goal of this course is to introduce you to the analysis of metabarcoding sequence data, with a specific focus on environmental DNA or eDNA, and make you comfortable to explore your own next-generation sequencing data. During this course, we will introduce the concept of metabarcoding and go through the different steps of a bioinformatic pipeline, from raw sequencing data to a count table with taxonomic assignments. At the end of this course, we hope you will be able to assemble paired-end sequences, demultiplex fastq sequencing files, filter sequences based on quality, cluster and denoise sequence data, build your own custom reference database, assign taxonomy to OTUs, further curate you data, and analyse the data in a statistically correct manner.
1. Introduction
Based on the excellent presentations at the start of today’s workshop, we can get an understanding of how broadly applicable eDNA metabarcoding is and how many research fields are using this technology to answer ecological questions.
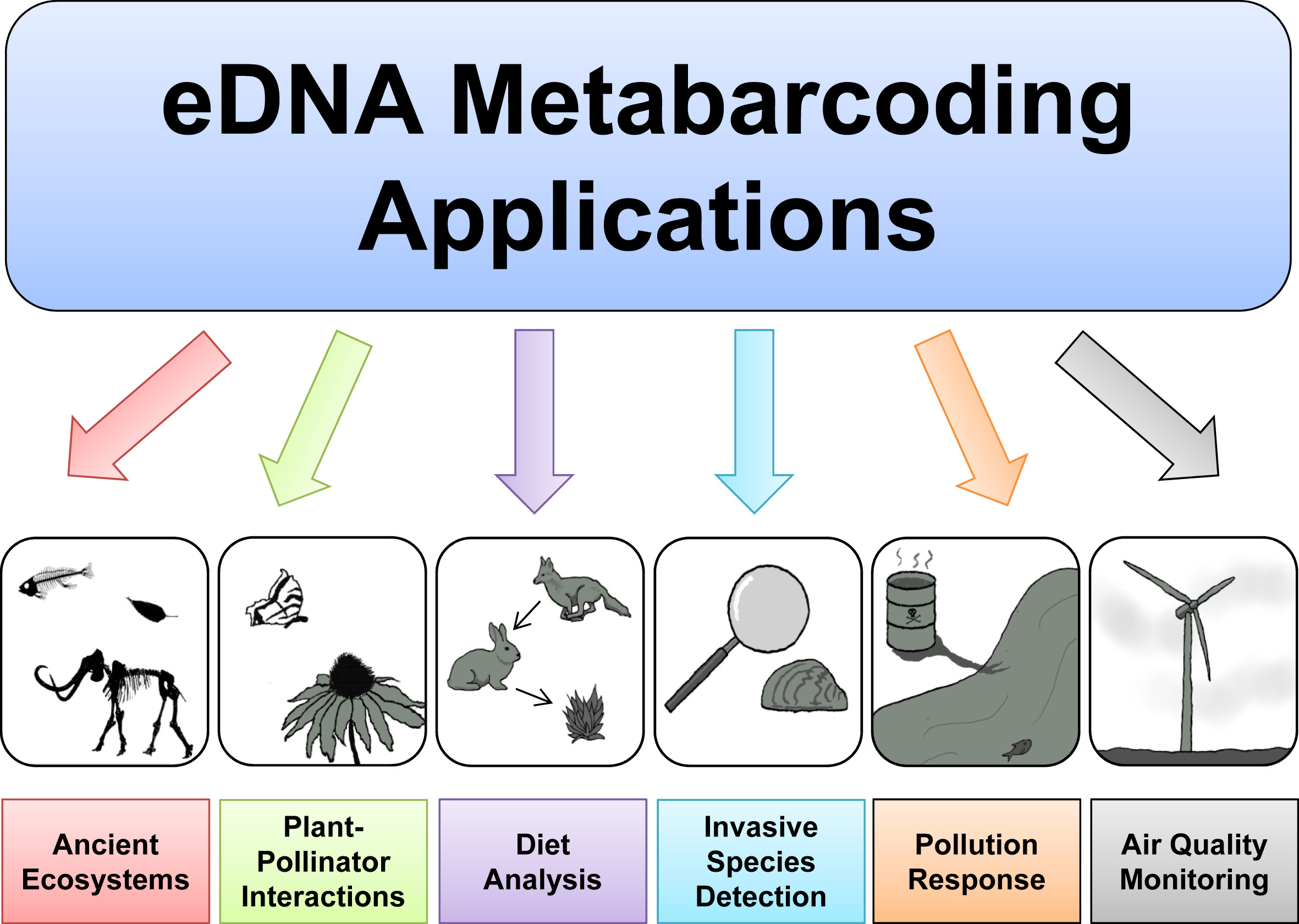
Before we get started with coding, we will introduce the dataset that we will be analysing today, some of the software programs available (including the ones we will use today), and the general structure of a bioinformatic pipeline to process metabarcoding data. Finally, we will go over the folder structure we suggest you use during the bioinformatic processing of metabarcoding data to help keep an overview of the files and ensure reproducibility of the bioinformatic pipeline.
2. Experimental setup
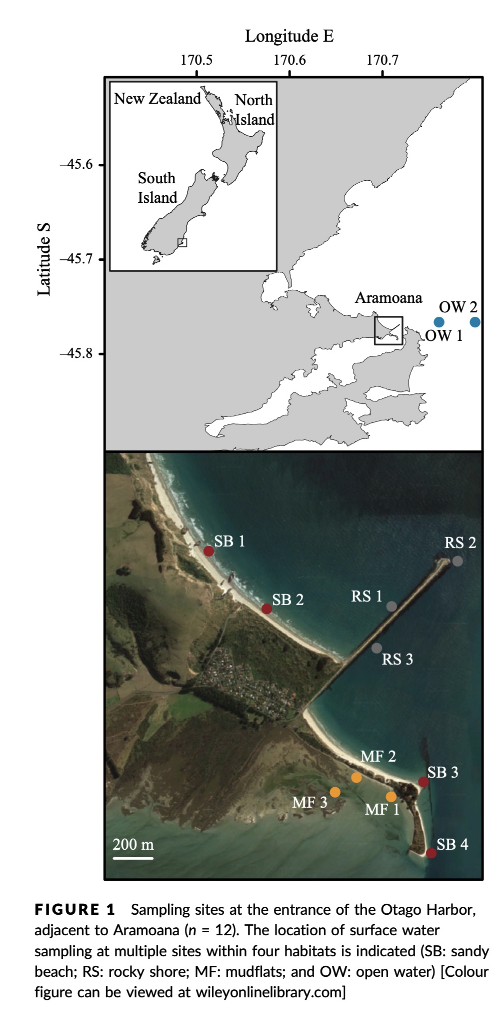
The data we will be analysing today is taken from an experiment we published in 2018 in Molecular Ecology Resources. In this experiment we wanted to determine the spatial resolution of aquatic eDNA in the marine environment. To investigate the spatial resolution, we visited a marine environment with high dynamic water movement where we knew from traditional monitoring that there were multiple habitats present on a small spatial scale containing different community assemblages. Our hypothesis was as follows, if the spatial resolution of aquatic eDNA is high, we would encounter distinct eDNA signals at each habitat resembling the residing community. However, if aquatic eDNA is transported through water movement, from currents and tidal waves, similar eDNA signals would be obtained across this small sampling area, which would be indicative of a low spatial resolution for aquatic eDNA. Our experiment consisted of five replicate water samples (2l) at each sampling point. Water was filtered using a vacuum pump and extracted using a standard Qiagen DNeasy Blood & Tissue Kit. All samples were amplified using three metabarcoding primer sets to target fish and crustacean diversity, specifically. We also used a generic COI primer set to amplify eukaryotic eDNA signals.
For today’s data analysis, we subsetted the data to only include two sampling sites and we’ll only be looking at the fish sequencing data. The reason for doing so, is to ensure all bioinformatic steps will take maximum a couple of minutes, rather than hours or days to complete the full analysis.

3. A note on software programs
The popularity of metabarcoding research, both in the bacterial and eukaryote kingdom, has resulted in the development of a myriad of software packages and pipelines. Furthermore, software packages are continuously updated with new features, as well as novel software programs being designed and published. We will be using several software programs and R packages during this workshop. However, we would like to stress that the pipeline used in the workshop is by no means better than other pipelines and we urge you to explore alternative software programs and trial them with your own data after this workshop.
Metabarcoding software programs can generally be split up into two categories, including:
- stand-alone programs that are self-contained: These programs usually incorporate novel functions or alterations on already-existing functions. Programs can be developed for a specific task within the bioinformatic pipeline or can execute multiple or all steps in the bioinformatic pipeline. Examples of such programs are: Mothur, USEARCH, VSEARCH, cutadapt, dada2, OBITools3.
- wrappers around existing software: These programs attempt to provide an ecosystem for the user to complete all steps within the bioinformatic pipeline without needing to reformat documents or change the coding language. Examples of such programs are: QIIME2 and JAMP.
Two notes of caution about software programs:
- While some software programs are only able to complete a specific aspect of the bioinformatic pipeline, others are able to handle all steps. It might therefore seem easier to use a program that can be used to run through the pipeline in one go. However, this could lead to a ‘black box’ situation and can limit you when other software programs might have additional options or better alternatives. It is, therefore, recommended to understand each step in the bioinformatic pipeline and how different software programs implement these steps. Something we hope you will take away from today’s eDNA workshop.
- One issue you will run into when implementing several software programs in your bioinformatic pipeline is that each program might change the structure of the files in a specific manner. These modifications might lead to incompatability issues when switching fto the next program. It is, therefore, important to understand how a specific program changes the structure of the sequencing file and learn how these modification can be reverted back (through simple python or bash scripts). We will talk a little bit more about file structure and conversion throughout the workshop.
Below, we can find a list of some of the software programs that are available for analysing metabarcoding data (those marked with a ‘*’ will be covered in this workshop):
-
FastQC* (webpage) is a program that assesses the quality of .fastq files.
-
MultiQC (webpage) is a program that combines multple FastQC reports into a single report.
-
Cutadapt* (webpage) is a program that finds and removes adapter sequences, primers, and other types of unwated sequences from high-throughput sequencing reads. We will use this program to demultiplex our data and remove the primer-binding regions.
-
USEARCH/VSEARCH* (USEARCH), and the open-source version (which we will use) (VSEARCH*), has many utilities for running metabarcoding analyses. Many of the most common pipelines use at least some of the tools from these packages.
-
CRABS* (webpage) is a software package developed by our team that lets you build and curate custom reference databases.
-
tombRaider* (webpage) is an algorithm developed by our team that identifies and removes artefacts from metabarcoding datasets.
-
Qiime2 is actually a collection of different programs and packages, all compiled into a single ecosystem so that all things can be worked together. On the Qiime2 webpage there is a wide range of tutorials and help.
-
Dada2 is a good program, run in the R language. There is good documentation and tutorials for this on its webpage. You can actually run Dada2 within Qiime, but there are a few more options when running it alone in R.
-
Mothur has long been a mainstay of metabarcoding analyses, and is worth a look
-
Another major program for metabarcoding is OBITools, which was one of the first to specialise in non-bacterial metabarcoding datasets. The latest version is built using the coding language GO and is currently in Beta. Check the main webpage to keep track of its development.
Additionally, there are several good R packages that can be used to analyse and graph results. Chief among these are Phyloseq, iNEXT.3D, ape, microbiome, indicspecies, vegan, and DECIPHER.
In a recent meta-analysis our lab conducted, we can see that DADA2 and USEARCH/VSEARCH are the most prominently used software programs in the metabarcoding research community.
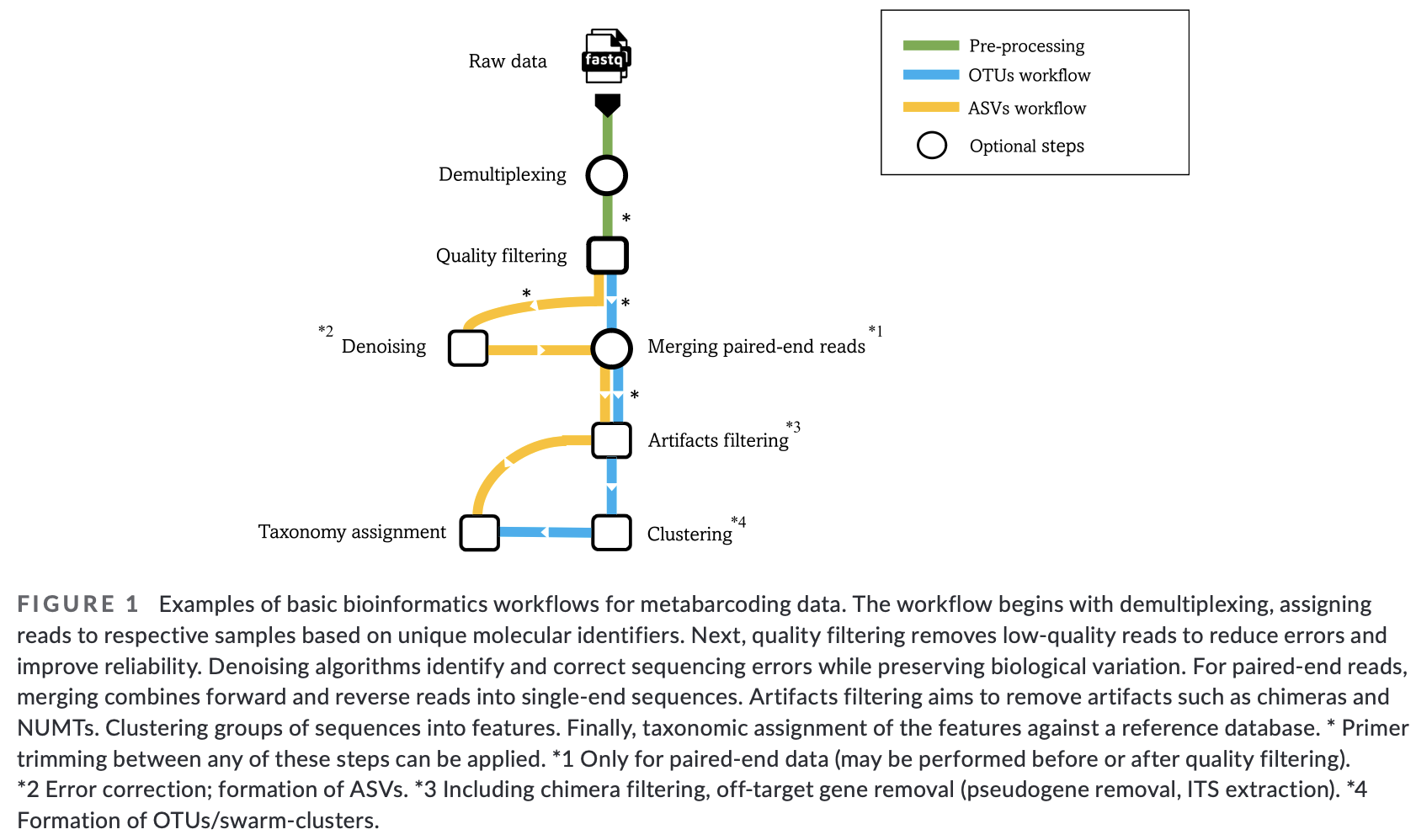
4. General overview of the bioinformatic pipeline
Although software options are numerous, each pipeline follows more or less the same basic steps and accomplishes these steps using similar tools. Before we get started with coding, let’s quickly go over the main steps of the pipeline to give you a clear overview of what we will cover today. For each of these steps, we will provide more information when we cover those sections during the workshop.
5. A note on scripts and reproducible code
One final topic we need to cover before we get started with coding is the use of scripts to execute code. During this workshop, we will create very small and simple scripts that contain the code used to process the sequencing data. While it is possible to run the commands without using scripts, it is good practice to use this method for the following three reasons:
- By using scripts to execute the code, there is a written record of how the data was processed. This will make it easier to remember how the data was processed in the future.
- While the sample data provided in this tutorial is small and computing steps take up a maximum of several minutes, processing large data sets can be time consuming. It is, therefore, recommended to process a small portion of the data first to test the code, modify the filenames once everything is set up, and run it on the full data set.
- Scripts can be reused in the future by changing file names, saving you time by not having to constantly write every line of code in the terminal. Minimising the amount needing to be written in the terminal will, also, minimise the risk of receiving error messages due to typo’s.
6. The folder structure
That is it for the intro, let’s get started with coding!
First, we will have a look at the folder structure that will be used during the workshop. For any project, it is important to organise your various files into subfolders. Through the course you will be navigating between folders to run your analyses. It may seem confusing or tedious at first, especially as there are only a few files we will be dealing with during this workshop. However, keep in mind that for many projects you can easily generate hundreds of files, so it is best to start with good practices from the beginning.
Let’s navigate to the edna folder in the ~/obss_2024/ directory. Running the ls -ltr command outputs the following subfolders:
- raw/
- input/
- output/
- meta/
- scripts/
- final/
- refdb/
- stats/
Most of these are currently empty, but by the end of the course they will be full of the results of your analysis.
So, let’s begin!
Key Points
Most metabarcoding pipelines include the same basic components
Frequency tables are key to metabarcoding analyses
Sequencing data preparation
Overview
Teaching: 20 min
Exercises: 10 minQuestions
What are the different primer design methods for metabarcoding?
How do we demultiplex and quality control metabarcoding data?
Objectives
Learn the different methods for QC of metabarcoding data
Sequence data preparation
1. Introduction
To start, let’s navigate to the raw sequencing file in the raw folder within the ~/obss_2024/edna/ directory and list the files in the subdirectory. To navigate between directories, we can use the cd command, which stands for change directory. It is always a good idea to check if the code executed as expected. In this case, we can verify the current working directory, i.e., the directory we navigated to with the cd command, by using the pwd command, which stands for print working directory.
$ cd ~/obss_2024/edna/raw/
$ pwd
/home/<username>/obss_2024/edna/raw
To list all the documents and subdirectories in our working directory, we can use the ls command. Additionally, we will specify the -ltr parameters to the ls command. The l parameter provides the list in long format that includes the permissions, which will come in handy when we are creating our scripts (but more on that later). The t parameter sorts the output by time and date, while the r parameter is printing the list in reverse order, i.e., the newest files will be printed at the bottom of the list. The reverse order using r is particularly handy when you are working with hundreds of files, as you don’t have to scroll up to find the files that were created using the latest executed command.
$ ls -ltr
-rw-r-----+ 1 jeuge18p nesi02659 124306274 Nov 20 08:46 FTP103_S1_L001_R1_001.fastq.gz
From the output, we can see that the sequencing file is currently zipped, i.e., file extension .gz. This is done to reduce file sizes and facilitate file transfer between the sequencing service and the customer. Most software will be able to read and use zipped files, so there is no need to unzip the file now. However, always check if software programs are capable of handling zipped files.
If we look at the left-hand side of the output, we can see the permissions of the files and directories in the list that was printed using the ls -ltr command. For our raw sequencing file, we have the r, w, and x permissions. Besides these 3 permissions, two additional ones also exist, namely d and @. Below is a list of what these permissions indicate.
d: directory - indicates that this is a folder or directory, rather than a file.r: read - specifies that we have access to read the file.w: write - specifies that we can write to the file.x: execute - specifies that we have permission to execute the file.@: extended - a novel symbol for MacOS indicating that the file has extended attributes (MacOS specific).
Finally, we can see that we have a single fastq file, the R1_001.fastq.gz file, indicating that the library was sequenced single-end. When you conduct paired-end sequencing, there will be a second file with an identical name, except for R2 rather than R1. Because of our single-end sequencing strategy, we do not need to merge or assemble paired reads (see Introduction for more information). Although we do not need to merge our reads, I have provided an example code below on how you might be able to merge paired-end sequencing data using VSEARCH. (DO NOT RUN THE CODE BELOW, AS IT WILL FAIL!)
vsearch --fastq_mergepairs FTP103_S1_L001_R1_001.fastq.gz --reverse FTP103_S1_L001_R2_001.fastq.gz --fastqout FTP103_merged.fastq --fastq_allowmergestagger
2. Quality control
The first thing we will do with our sequencing file is to look at the quality and make sure everything is according to the sequencing parameters that were chosen. The program we will be using for this is FastQC. However, since you’ve already encountered this software program in a previous day, we’ll just focus on the output for now.
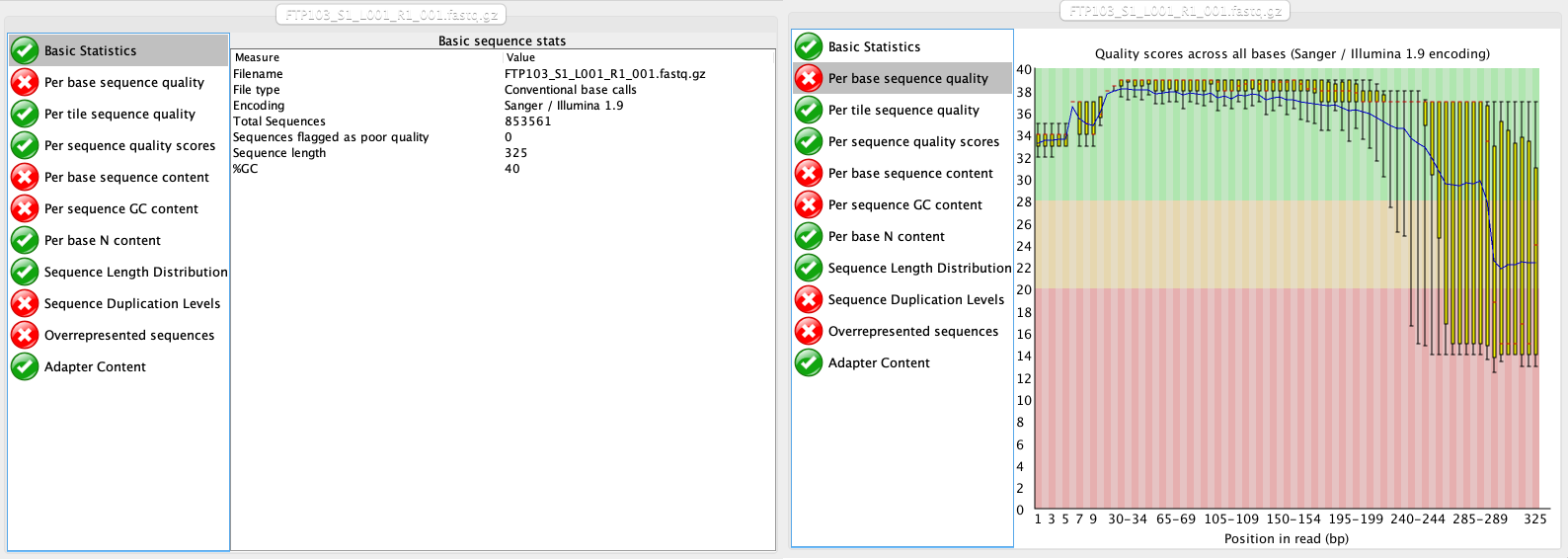
At this stage, we will be looking at the tabs Basic Statistics and Per base sequence quality. From this report, we can see that:
- We have a total of 853,561 sequences.
- The sequence length is 325 bp, identical to the cycle number of the specified sequencing kit.
- Sequence quality is dropping significantly at the end, which is typical for Illumina sequencing data.
3. Demultiplexing
3.1 Introduction to demultiplexing
Given that we only have a single sequence file containing all reads for multiple samples, the first step in our pipeline will be to assign each sequence to a sample, a process referred to as demultiplexing. To understand this step in the pipeline, we will need to provide some background on library preparation methods.
As of the time of writing this workshop, PCR amplification is the main method to generate eDNA metabarcoding libraries. To achieve species detection from environmental or bulk specimen samples, PCR amplification is carried out with primers designed to target a taxonomically informative marker within a taxonomic group. Furthermore, next generation sequencing data generates millions of sequences in a single sequencing run. Therefore, multiple samples can be combined. To achieve this, a unique sequence combination is added to the amplicons of each sample. This unique sequence combination is usually between 6 and 8 base pairs long and is referred to as ‘tag’, ‘index’, or ‘barcode’. Besides amplifying the DNA and adding the unique barcodes, sequencing primers and adapters need to be incorporated into the library to allow the DNA to be sequenced by the sequencing machine. How barcodes, sequencing primers, and adapters are added, and in which order they are added to your sequences will influence the data files you will receive from the sequencing service.
For example, traditionally prepared Illumina libraries (ligation and two-step PCR) are constructed in the following manner (the colours mentioned refer to the figure below):
- Forward Illumina adapter (red)
- Forward barcode sequence (darkgreen)
- Forward Illumina sequencing primer (orange)
- Amplicon (including primers)
- Reverse Illumina sequencing primer (lightblue)
- Reverse barcode sequence (lightgreen)
- Reverse Illumina adapter (darkblue)
This construction allows the Illumina software to identify the barcode sequences, which will result in the generation of a separate sequence file (or two for paired-end sequencing data) for each sample. This process is referred to as demultiplexing and is performed by the Illumina software.
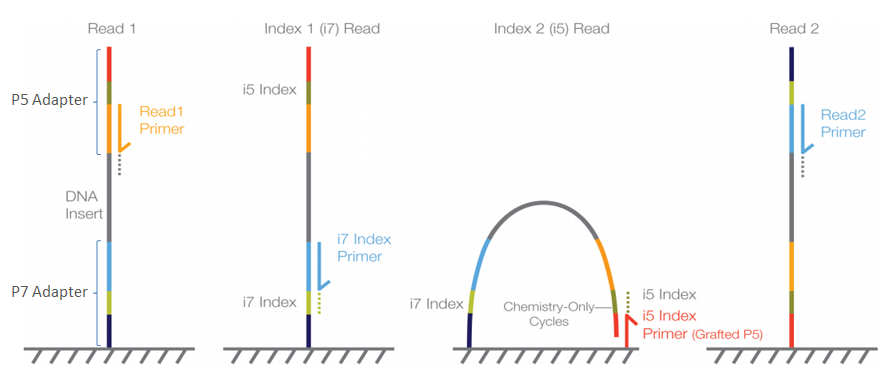
When libraries are prepared using the single-step PCR method, the libraries are constructed slightly different (the colours mentioned refer to the setup described in the two-step approach):
- Forward Illumina adapter (red)
- Forward Illumina sequencing primer (orange)
- Forward barcode sequence (darkgreen)
- Amplicon (including primers)
- Reverse barcode sequence (lightgreen)
- Reverse Illumina sequencing primer (lightblue)
- Reverse Illumina adapter (darkblue)
In this construction, the barcode sequences are not located in between the Illumina adapter and Illumina sequencing primer, but are said to be ‘inline’. These inline barcode sequences cannot be identified by the Illumina software, since it does not know when the barcode ends and the primer sequence of the amplicon starts. Therefore, this will result in a single sequence file (or two for paired-end sequencing data) containing the data for all samples together. The demultiplexing step, assigning sequences to a sample, will have to be incorporated into the bioinformatic pipeline, something we will cover now in the workshop.
For more information on Illumina sequencing, you can watch the following 5-minute youtube video (EMP). Notice that the ‘Sample Prep’ method described in the video follows the two-step PCR approach.
3.2 Assigning sequences to corresponding samples
During library preparation for our sample data, each sample was assigned a specific barcode combination, with at least 3 basepair differences between each barcode used in the library. By searching for these specific sequences, which are situated at the very beginning and very end of each sequence, we can assign each sequence to the corresponding sample. We will be using cutadapt for demultiplexing our dataset today, as it is the fastest option. Cutadapt requires an additional metadata text file in fasta format. This text file contains information about the sample name and the associated barcode and primer sequences.
Let’s have a look at the metadata file, which can be found in the meta subfolder.
$ cd ../meta/
$ ls -ltr
-rw-r-----+ 1 jeuge18p nesi02659 1139 Nov 20 08:46 sample_metadata.fasta
To investigate the first couple of lines of a text file (unzipped), we can use the head command. The number of the -n parameter specifies the number of lines that will be printed.
$ head -n 10 sample_metadata.fasta
>AM1
GAAGAGGACCCTATGGAGCTTTAGAC;min_overlap=26...AGTTACYHTAGGGATAACAGCGCGACGCTA;min_overlap=30
>AM2
GAAGAGGACCCTATGGAGCTTTAGAC;min_overlap=26...AGTTACYHTAGGGATAACAGCGCGAGTAGA;min_overlap=30
>AM3
GAAGAGGACCCTATGGAGCTTTAGAC;min_overlap=26...AGTTACYHTAGGGATAACAGCGCGAGTCAT;min_overlap=30
>AM4
GAAGAGGACCCTATGGAGCTTTAGAC;min_overlap=26...AGTTACYHTAGGGATAACAGCGCGATAGAT;min_overlap=30
>AM5
GAAGAGGACCCTATGGAGCTTTAGAC;min_overlap=26...AGTTACYHTAGGGATAACAGCGCGATCTGT;min_overlap=30
From the output, we can see that this is a simple two-line fasta format, with the header line containing the sample name info indicated by ”>” and the barcode/primer info on the following line. We can also see that the forward and reverse barcode/primer sequences are split by “…“ and that we specify a minimum overlap between the sequence found in the data file and the metadata file to be the full length of the barcode/primer information.
Note that by specifying the barcode + primer region, cutadapt will automatically identify which sequence belongs to which sample (barcode) and remove all artificial parts of our sequence (barcode + primer) to only retain our amplicon sequence. If you receive already-demultiplexed data from the sequencing service, be aware that you most likely need to remove the primer regions yourself. This can be done using the cutadapt code below as well.
Count the number of samples in the metadata file
before we get started with our first script, let’s first determine how many samples are included in our experiment.
Hint 1: think about the file structure of the metadata file.
Hint 2: a similar line of code can be written to count the number of sequences in a fasta file (which you might have come across before during OBSS2024)
Solution
grep -c "^>" sample_metadata.fasta
grepis a command that searches for strings that match a pattern-cis the parameter indicating to count the number of times we find the pattern"^>"is the string we need to match, in this case>. The^indicates that the string needs to occur at the beginning of the line.
Let’s now write our first script to demultiplex our sequence data. First, we’ll navigate to the scripts folder. When listing the files in this folder, we see that a script has already been created, so let’s see what it contains.
$ cd ../scripts/
$ ls -ltr
-rwxr-x---+ 1 jeuge18p nesi02659 189 Nov 20 08:46 moduleload
To read a text file in its entirity, we can use the nano command, which will open a new window.
$ nano moduleload
#!/bin/bash
module load CRABS/1.0.6
module load FastQC/0.12.1
module load cutadapt/4.4-gimkl-2022a-Python-3.11.3
module load VSEARCH/2.21.1-GCC-11.3.0
module load BLAST/2.13.0-GCC-11.3.0
When opening this file, we can see that the script starts with what is called a shebang #!. This sign lets the computer know how to interpret the script. The following lines are specific to NeSI, where we will load the necessary software programs to run our scripts. We will, therefore, add this script to all subsequent scripts to make sure the correct software is available on NeSI.
To exit the script, we can press ctr + X, which will bring us back to the terminal window.
3.3 Our first script
To create a new script to demultiplex our data, we can type the following:
$ nano demux
This command will open the file demux in the nano text editor in the terminal. In here, we can type the following commands:
$ #!/bin/bash
$
$ source moduleload
$
$ cd ../raw/
$
$ cutadapt FTP103_S1_L001_R1_001.fastq.gz -g file:../meta/sample_metadata.fasta -o ../input/{name}.fastq --discard-untrimmed --no-indels -e 0 --cores=0
We can exit out of the text editor by pressing ctr+X, then type y to save the file, and press enter to save the file as demux. When we list the files in the scripts folder, we can see that this file is now created, but that it is not yet executable (no x in file description).
$ ls -ltr
-rwxr-x---+ 1 jeuge18p nesi02659 189 Nov 20 08:46 moduleload
-rw-rw----+ 1 jeuge18p nesi02659 194 Nov 20 09:09 demux
To make the script executable, we can use the following code:
$ chmod +x demux
When we now list the files in our scripts folder again, we see that the parameters have changed:
$ ls -ltr
-rwxr-x---+ 1 jeuge18p nesi02659 189 Nov 20 08:46 moduleload
-rwxrwx--x+ 1 jeuge18p nesi02659 194 Nov 20 09:09 demux
To run the script, we can type:
$ ./demux
Once this script has been executed, it will generate quite a bit of output in the terminal. The most important information, however, can be found at the top, indicating how many sequences could be assigned. The remainder of the output is detailed information for each sample.
This is cutadapt 4.4 with Python 3.11.3
Command line parameters: FTP103_S1_L001_R1_001.fastq.gz -g file:../meta/sample_metadata.fasta -o ../input/{name}.fastq --discard-untrimmed --no-indels -e 0 --cores=0
Processing single-end reads on 70 cores ...
Done 00:00:08 853,561 reads @ 9.5 µs/read; 6.33 M reads/minute
Finished in 8.136 s (9.532 µs/read; 6.29 M reads/minute).
=== Summary ===
Total reads processed: 853,561
Reads with adapters: 128,874 (15.1%)
== Read fate breakdown ==
Reads discarded as untrimmed: 724,687 (84.9%)
Reads written (passing filters): 128,874 (15.1%)
Total basepairs processed: 277,407,325 bp
Total written (filtered): 26,075,782 bp (9.4%)
If successfully completed, the script should’ve generated a single fastq file for each sample in the metadata file and placed these files in the input directory. Let’s see if the files have been created using the ls command.
$ ls -ltr ../input/
-rw-rw----+ 1 jeuge18p nesi02659 5404945 Nov 20 09:10 AM1.fastq
-rw-rw----+ 1 jeuge18p nesi02659 4906020 Nov 20 09:10 AM2.fastq
-rw-rw----+ 1 jeuge18p nesi02659 3944614 Nov 20 09:10 AM3.fastq
-rw-rw----+ 1 jeuge18p nesi02659 5916544 Nov 20 09:10 AM4.fastq
-rw-rw----+ 1 jeuge18p nesi02659 5214931 Nov 20 09:10 AM5.fastq
-rw-rw----+ 1 jeuge18p nesi02659 4067965 Nov 20 09:10 AM6.fastq
-rw-rw----+ 1 jeuge18p nesi02659 4841565 Nov 20 09:10 AS2.fastq
-rw-rw----+ 1 jeuge18p nesi02659 9788430 Nov 20 09:10 AS3.fastq
-rw-rw----+ 1 jeuge18p nesi02659 5290868 Nov 20 09:10 AS4.fastq
-rw-rw----+ 1 jeuge18p nesi02659 5660255 Nov 20 09:10 AS5.fastq
-rw-rw----+ 1 jeuge18p nesi02659 4524100 Nov 20 09:10 AS6.fastq
-rw-rw----+ 1 jeuge18p nesi02659 461 Nov 20 09:10 ASN.fastq
Count the number of demultiplexed files
We know from the metadata file that we will be analysing 12 files. While with this low number we can quickly count the number of items in the list, counting files in this manner becomes more difficult for larger projects. How would you calculate the number of files that were created by cutadapt?
Hint 1: we know the directory the files were created in and how to create the list of files.
Hint 2: make use of piping and counting functions in bash
Solution
ls -1 ../input/ | wc -l
|is a pipe that parses the output from one command into the input of the nextwcis a function to count items (word count)-lis a parameter that specifies that what should be counted are lines, not words.
Count the number of demultiplexed sequences
cutadapt reported that 128,874 sequences were demultiplexed. How would you count the total number of sequences across the 12 demultiplexed files?
Hint 1: think about the file structure of fastq files
Hint 2: make use of piping and counting functions in bash
Solution
cat ../input/*.fastq | wc -l | awk '{print $1 / 4}'
catis a function to combine all 12 fastq files in theinputdirectory|is a pipe that parses the output from one command into the input of the nextwcis a function to count items (word count)-lis a parameter that specifies that what should be counted are lines, not wordsawkis a very powerful computer language to parse text{print $1 / 4}is the parameter passed toawkto print the number of lines found bywcand divides this number by 4, due to the file structure of a fastq file.
4 Renaming sequence headers to incorporate sample names
Before continuing, let’s have a look at the file structure of the demultiplexed fastq files using the head command.
$ head -n 12 ../input/AM1.fastq
@M02181:93:000000000-D20P3:1:1101:15402:1843 1:N:0:1
GACAGGTAGACCCAACTACAAAGGTCCCCCAATAAGAGGACAAACCAGAGGGACTACTACCCCCATGTCTTTGGTTGGGGCGACCGCGGGGGAAGAAGTAACCCCCACGTGGAACGGGAGCACAACTCCTTGAACTCAGGGCCACAGCTCTAAGAAACAAAATTTTTGACCTTAAGATCCGGCAATGCCGATCAACGGACCG
+
2222E2GFFCFFCEAEF3FFBD32FFEFEEE1FFD55BA1AAEEFFE11AA??FEBGHEGFFGE/F3GFGHH43FHDGCGEC?B@CCCC?C??DDFBF:CFGHGFGFD?E?E?FFDBD.-A.A.BAFFFFFFBBBFFBB/.AADEF?EFFFF///9BBFE..BFFFF;.;FFFB///BBFA-@=AFFFF;-BADBB?-ABD;
@M02181:93:000000000-D20P3:1:1101:16533:1866 1:N:0:1
TTTAGAACAGACCATGTCAGCTACCCCCTTAAACAAGTAGTAATTATTGAACCCCTGTTCCCCTGTCTTTGGTTGGGGCGACCACGGGGAAGAAAAAAACCCCCACGTGGACTGGGAGCACCTTACTCCTACAACTACGAGCCACAGCTCTAATGCGCAGAATTTCTGACCATAAGATCCGGCAAAGCCGATCAACGGACCG
+
GGHCFFBFBB3BA2F5FA55BF5GGEGEEHF55B5ABFB5F55FFBGGB55GEEEFEGFFEFGC1FEFFGBBGH?/??EC/>E/FE???//DFFFFF?CDDFFFCFCF...>DAA...<.<CGHFFGGGH0CB0CFFF?..9@FB.9AFFGFBB09AAB?-/;BFFGB/9F/BFBB/BFFF;BCAF/BAA-@B?/B.-.@DA
@M02181:93:000000000-D20P3:1:1101:18101:1874 1:N:0:1
ACTAGACAACCCACGTCAAACACCCTTACTCTGCTAGGAGAGAACATTGTGGCCCTTGTCTCACCTGTCTTCGGTTGGGGCGACCGCGGAGGACAAAAAGCCTCCATGTGGACTGAAGTAACTATCCTTCACAGCTCAGAGCCGCGGCTCTACGCGACAGAAATTCTGACCAAAAATGATCCGGCACATGCCGATTAACGGAACA
+
BFGBA4F52AEA2A2B2225B2AEEGHFFHHF5BD551331113B3FGDGB??FEGG2FEEGBFFGFGCGHE11FE??//>//<</<///<</CFFB0..<F1FE0G0D000<<AC0<000<DDBC:GGG0C00::C0/00:;-@--9@AFBB.---9--///;F9F//;B/....;//9BBA-9-;.BBBB>-;9A/BAA.999
We can see that the sequences within each file do not contain any information to which sample they belong to. Currently, the sequences still have the initial Illumina sequence headers. To make things a bit easier for us, it would be best if the sequences themselves would contain the sample info, rather than having a file per sample. This would enable us to merge all files and run the bioinformatic code on a single file, rather than having to write loops to execute the code on each file separately, which can be a bit more daunting and complex. Luckily, we can rename the sequence headers using VSEARCH, after which we can combine all files using the cat command.
Let’s write our second script!
$ nano rename
$ #!/bin/bash
$
$ source moduleload
$
$ cd ../input/
$
$ for fq in *.fastq; do vsearch --fastq_filter $fq --relabel $fq. --fastqout rl_$fq; done
$
$ cat rl* > ../output/relabel.fastq
$ chmod +x rename
$ ./rename
Once executed, the script will first rename the sequence headers of each file and store them in a new file per sample. Lastly, we combine all relabeled files using the cat command. This combined file can be found in the output folder. Let’s see if it is there and how the sequence headers have been altered:
$ ls -ltr ../output/
-rw-rw----+ 1 jeuge18p nesi02659 54738505 Nov 20 09:19 relabel.fastq
head -n 12 ../output/relabel.fastq
@AM1.1
GACAGGTAGACCCAACTACAAAGGTCCCCCAATAAGAGGACAAACCAGAGGGACTACTACCCCCATGTCTTTGGTTGGGGCGACCGCGGGGGAAGAAGTAACCCCCACGTGGAACGGGAGCACAACTCCTTGAACTCAGGGCCACAGCTCTAAGAAACAAAATTTTTGACCTTAAGATCCGGCAATGCCGATCAACGGACCG
+
2222E2GFFCFFCEAEF3FFBD32FFEFEEE1FFD55BA1AAEEFFE11AA??FEBGHEGFFGE/F3GFGHH43FHDGCGEC?B@CCCC?C??DDFBF:CFGHGFGFD?E?E?FFDBD.-A.A.BAFFFFFFBBBFFBB/.AADEF?EFFFF///9BBFE..BFFFF;.;FFFB///BBFA-@=AFFFF;-BADBB?-ABD;
@AM1.2
TTTAGAACAGACCATGTCAGCTACCCCCTTAAACAAGTAGTAATTATTGAACCCCTGTTCCCCTGTCTTTGGTTGGGGCGACCACGGGGAAGAAAAAAACCCCCACGTGGACTGGGAGCACCTTACTCCTACAACTACGAGCCACAGCTCTAATGCGCAGAATTTCTGACCATAAGATCCGGCAAAGCCGATCAACGGACCG
+
GGHCFFBFBB3BA2F5FA55BF5GGEGEEHF55B5ABFB5F55FFBGGB55GEEEFEGFFEFGC1FEFFGBBGH?/??EC/>E/FE???//DFFFFF?CDDFFFCFCF...>DAA...<.<CGHFFGGGH0CB0CFFF?..9@FB.9AFFGFBB09AAB?-/;BFFGB/9F/BFBB/BFFF;BCAF/BAA-@B?/B.-.@DA
@AM1.3
ACTAGACAACCCACGTCAAACACCCTTACTCTGCTAGGAGAGAACATTGTGGCCCTTGTCTCACCTGTCTTCGGTTGGGGCGACCGCGGAGGACAAAAAGCCTCCATGTGGACTGAAGTAACTATCCTTCACAGCTCAGAGCCGCGGCTCTACGCGACAGAAATTCTGACCAAAAATGATCCGGCACATGCCGATTAACGGAACA
+
BFGBA4F52AEA2A2B2225B2AEEGHFFHHF5BD551331113B3FGDGB??FEGG2FEEGBFFGFGCGHE11FE??//>//<</<///<</CFFB0..<F1FE0G0D000<<AC0<000<DDBC:GGG0C00::C0/00:;-@--9@AFBB.---9--///;F9F//;B/....;//9BBA-9-;.BBBB>-;9A/BAA.999
Finally, to check if no sequences were lost during renaming, we can use the wc command.
wc -l ../output/relabel.fastq | awk '{print $1 / 4}'
128874
Key Points
First key point. Brief Answer to questions. (FIXME)
Bioinformatic analysis
Overview
Teaching: 30 min
Exercises: 60 minQuestions
What parameters to use to filter metabarcoding data on quality?
Is it better to denoise or cluster sequence data?
How are chimeric sequences formed?
Objectives
Learn to filter sequences on quality using the VSEARCH pipeline
Learn to denoise sequences into ZOTUs with the VSEARCH pipeline
Learn to remove chimeric sequences from the dataset
Generate a count table
Bioinformatic analysis
1. Quality filtering
1.1 Introduction
During the bioinformatic pipeline, it is critical to only retain high-quality reads to reduce the abundance and impact of spurious sequences. There is an intrinsic error rate to all polymerases used during PCR amplification, as well as sequencing technologies. For example, the most frequently used polymerase during PCR is Taq, though lacks 3’ to 5’ exonuclease proofreading activity, resulting in relatively low replication fidelity. These errors will generate some portion of sequences that vary from their biological origin sequence. Such reads can substantially inflate metrics such as alpha diversity, especially in a denoising approach (more on this later). While it is near-impossible to remove all of these sequences bioinformatically, especially PCR errors, we will attempt to remove erroneous reads by filtering on base calling quality scores (the fourth line of a sequence record in a .fastq file).
For quality filtering, we will discard all sequences that do not adhere to a specific set of rules. We will be using the program VSEARCH (EMP) for this step. Quality filtering parameters are not standardized, but rather specific for each library. For our tutorial data, we will filter out all sequences that do not adhere to a minimum and maximum length, have unassigned base calls (‘N’), and have a higher expected error than 1. Once we have filtered our data, we can check the quality with FastQC and compare it to the raw sequencing file. As this is the last step in our bioinformatic pipeline where quality scores are essential, we will export our output files both in .fastq and .fasta format.
1.2 Filtering sequence data
$ nano qual
$ #!/bin/bash
$
$ source moduleload
$
$ cd ../output/
$
$ vsearch --fastq_filter relabel.fastq --fastq_maxee 1.0 --fastq_maxlen 230 --fastq_minlen 150 --fastq_maxns 0 --fastaout filtered.fasta --fastqout filtered.fastq
$ chmod +x qual
$ ./qual
vsearch v2.21.1_linux_x86_64, 503.8GB RAM, 72 cores
https://github.com/torognes/vsearch
Reading input file 100%
123537 sequences kept (of which 0 truncated), 5337 sequences discarded.
After quality filtering, we have 123,537 sequences left over in the fastq and fasta files. Before we continue with the pipeline, let’s check if the quality filtering was successful using FastQC.
nano qualcheck
#!/bin/bash
source moduleload
cd ../output/
fastqc filtered.fastq -o ../meta/
$ chmod +x qualcheck
$ ./qualcheck
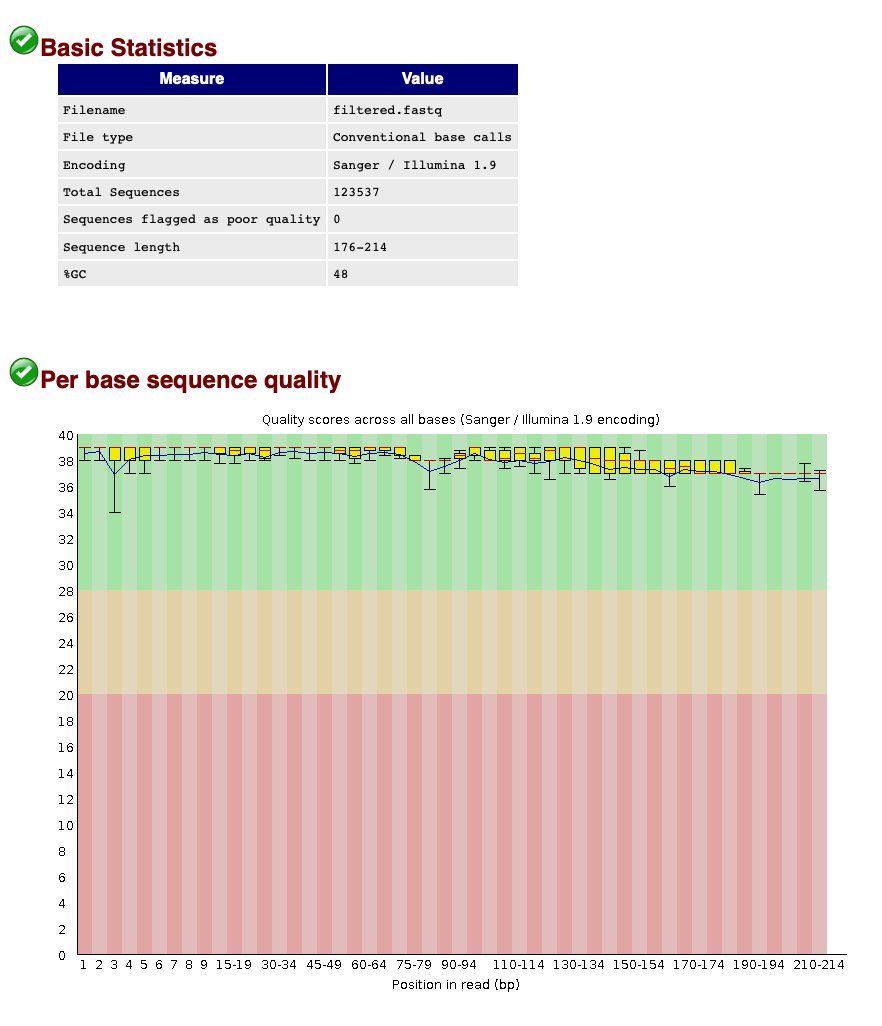
When we now look at the Basic Statistics and Per base sequence quality tabs, we can see that:
- Our total number of sequences has been reduced to 123,537, similar to the number specified by our
qualscript. - The sequence length ranges from 176-214, according to amplicon length and within the range of the parameters chosen in the
qualscript. - The sequence quality is high throughout the amplicon length (within the green zone).
We can, therefore, conclude that quality filtering was successful and we can now move to the clustering step of the pipeline.
Count the number of quality filtered sequences for each sample
Since we are working with a single file, VSEARCH reports the total number of sequences that were retained and discarded. How would we calculate the number of sequences that were retained for each sample?
Hint 1: think about the sequence header structure
Hint 2: use the solution in the previous exercise where we calculated the number of sequences across multiple files.
Solution
grep "^>" ../output/filtered.fasta | cut -d '.' -f 1 | sort | uniq -c
2. Dereplication
Once we have verified that the reads passing quality filtering are of high quality and of a length similar to the expected amplicon size, we need to dereplicate the data into unique sequences. Since metabarcoding is based on a PCR amplification method, the same DNA molecule will have been copied thousands or millions of times and, therefore, sequenced multiple times. In order to reduce the file size and computational cost, it is convenient to work with unique sequences and keep track of how many times each unique sequence was found in our data set. This dereplication step is also the reason why we combined all samples into a single file, as the same amplicon (or species if you will) can be found across different samples.
nano derep
#!/bin/bash
source moduleload
cd ../output/
vsearch --derep_fulllength filtered.fasta --relabel uniq. --sizeout --output uniques.fasta
chmod +x derep
./derep
vsearch v2.21.1_linux_x86_64, 503.8GB RAM, 72 cores
https://github.com/torognes/vsearch
Dereplicating file filtered.fasta 100%
25083932 nt in 123537 seqs, min 176, max 214, avg 203
Sorting 100%
6645 unique sequences, avg cluster 18.6, median 1, max 19554
Writing FASTA output file 100%
To determine if everything executed properly, we can check the uniques.fasta file using the head command.
$ head -n 12 ../output/uniques.fasta
>uniq.1;size=19554
TTTAGAACAGACCATGTCAGCTACCCCCTTAAACAAGTAGTAATTATTGAACCCCTGTTCCCCTGTCTTTGGTTGGGGCG
ACCACGGGGAAGAAAAAAACCCCCACGTGGACTGGGAGCACCTTACTCCTACAACTACGAGCCACAGCTCTAATGCGCAG
AATTTCTGACCATAAGATCCGGCAAAGCCGATCAACGGACCG
>uniq.2;size=14321
ACTAAGGCATATTGTGTCAAATAACCCTAAAACAAAGGACTGAACTGAACAAACCATGCCCCTCTGTCTTAGGTTGGGGC
GACCCCGAGGAAACAAAAAACCCACGAGTGGAATGGGAGCACTGACCTCCTACAACCAAGAGCTGCAGCTCTAACTAATA
GAATTTCTAACCAATAATGATCCGGCAAAGCCGATTAACGAACCA
>uniq.3;size=8743
ACCAAAACAGCTCCCGTTAAAAAGGCCTAGATAAAGACCTATAACTTTCAATTCCCCTGTTTCAATGTCTTTGGTTGGGG
CGACCGCGGAGTACTACCAAACCTCCATGTGGACCGAAAGAACATCTTTTATAGCTCCGAGTGACAACTCTAAGTAACAG
AACATCTGACCAGTATGATCCGGCATAGCCGATCAACGAACCG
Study Question 1
What do you think the following parameters are doing?
--sizeout--relabel uniq.Solution
--sizeout: this will print the number of times a sequence has occurred in the header information. You can use theheadcommand on the sorted file to check how many times the most abundant sequence was encountered in our sequencing data.
--relabel uniq.: this parameter will relabel the header information touniq.followed by an ascending number, depending on how many different sequences have been encountered.
Count the number of singleton sequences
VSEARCH has reported the total number of unique sequences. How would you calculate the number of singleton sequences in our file?
Hint 1: think about the sequence header structure
Hint 2: make sure to not count sequences where the number starts with 1, e.g., 10.
Solution
grep -c "size=1\b" ../output/uniques.fasta
Print the most and least abundant sequences
How would you determine the number of times the 10 most and 10 least abundant unique sequences were observed?
Hint 1: keep in mind the order of the sequences in the file.
Solution
awk '/^>uniq/ {a[i++]=$0} END {for (j=i-10; j<i; j++) print a[j]; for (k=0; k<10; k++) print a[k]}' ../output/uniques.fasta
3. Denoising
3.1 Introduction
Now that our data set is filtered and unique sequences have been retrieved, we are ready for the next step in the bioinformatic pipeline, i.e., looking for biologically meaningful or biologically correct sequences. Two approaches to achieve this goal exist, including denoising and clustering. There is still an ongoing debate on what the best approach is to obtain these biologically meaningful sequences. For more information, these are two good papers to start with: Brandt et al., 2021; Antich et al., 2021. For this workshop, we won’t be discussing this topic in much detail, but this is the basic idea…
When clustering the dataset, OTUs (Opterational Taxonomic Units) will be generated by combining sequences that are similar to a set percentage level (traditionally 97%), with the most abundant sequence identified as the true sequence. When clustering at 97%, this means that if a sequence is more than 3% different than the generated OTU, a second OTU will be generated. The concept of OTU clustering was introduced in the 1960s and has been debated since. Clustering the dataset is usually used for identifying species in metabarcoding data.
Denoising, on the other hand, attempts to identify all correct biological sequences through an algorithm, which is visualised in the figure below. In short, denoising will cluster the data with a 100% threshold and tries to identify errors based on abundance differences. The retained sequences are called ZOTU (Zero-radius Operation Taxonomic Unit) or ASVs (Amplicon Sequence Variants). Denoising the dataset is usually used for identifying intraspecific variation in metabarcoding data. A schematic of both approaches can be found below.
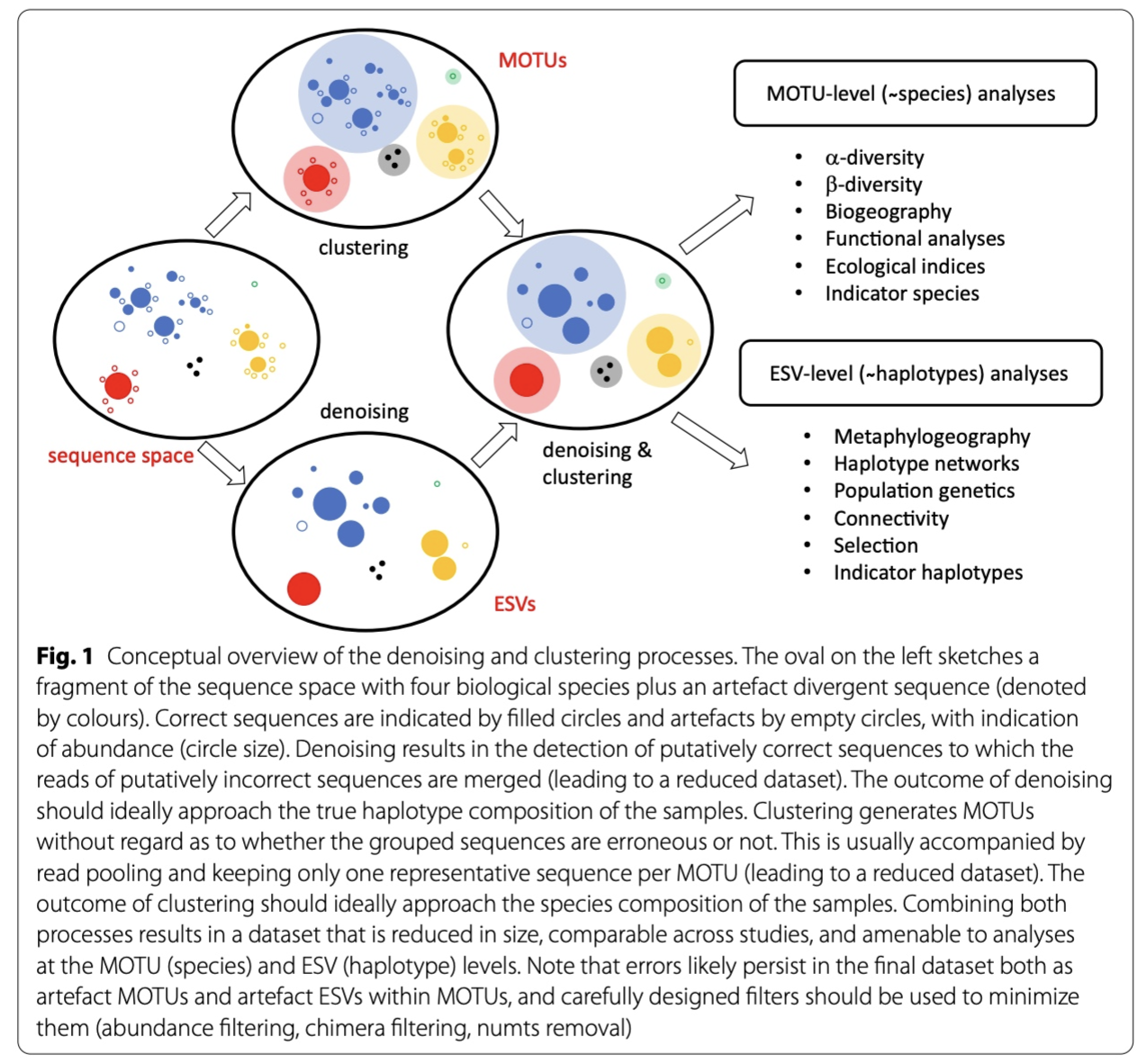
This difference in approach may seem small but has a very big impact on your final dataset!
When you denoise the dataset, it is expected that one species may have more than one ZOTU. This means that diversity estimates will be highly inflated. Luckily, we have recently developed an algorithm to counteract this issue (but more on this later). When clustering the dataset, on the other hand, it is expected that an OTU may have more than one species assigned to it, meaning that you may lose some correct biological sequences that are present in your data by merging species with barcodes more similar than 97%. In other words, you will miss out on differentiating closely related species and intraspecific variation.
For this workshop, we will follow the denoising approach, as it is favoured in recent years. Plus with novel data curation methods, e.g., tombRaider (introduced later in this workshop), the issue of diversity inflation has been largely resolved. However, clustering is still a valid option. So, feel free to explore this approach for your own data set if you prefer.

3.2 Denoising the data
For denoising, we will use the unoise3 algorithm as implemented in VSEARCH. Since denoising is based on read abundance of the unique sequences, we can specify the --sizein parameter. The minimum abundance threshold for a true denoised read is defaulted to 8 reads, as specified by the unoise3 algorithm developer. However, more recent research by Bokulich et al., 2013, identified a minimum abundance threshold to be more appropriate. Hence, we will set the --minsize parameter to 0.0001%, which in our case is ~12 reads.
nano denoise
#!/bin/bash
source moduleload
cd ../output/
vsearch --cluster_unoise uniques.fasta --sizein --minsize 12 --sizeout --relabel denoised. --centroids denoised.fasta
chmod +x denoise
./denoise
vsearch v2.21.1_linux_x86_64, 503.8GB RAM, 72 cores
https://github.com/torognes/vsearch
Reading file uniques.fasta 100%
46023 nt in 227 seqs, min 189, max 213, avg 203
minsize 12: 6418 sequences discarded.
Masking 100%
Sorting by abundance 100%
Counting k-mers 100%
Clustering 100%
Sorting clusters 100%
Writing clusters 100%
Clusters: 49 Size min 12, max 24181, avg 4.6
Singletons: 0, 0.0% of seqs, 0.0% of clusters
From the output, we can see that we have generated a list of 49 ZOTUs. Additionally, we can see that 6,418 out of 6,645 unique sequences were discarded, as they didn’t reach the minimum abundance threshold of 12 sequences.
4. Chimera removal
The second to last step in our bioinformatic pipeline is to remove chimeric sequences. Amplicon sequencing has the potential to generate chimeric reads, which can cause spurious inference of biological variation. Chimeric amplicons form when an incomplete DNA strand anneals to a different template and primes synthesis of a new template derived from two different biological sequences, or in other words chimeras are artefact sequences formed by two or more biological sequences incorrectly joined together. More information can be found within this paper and a simple illustration can be found below.
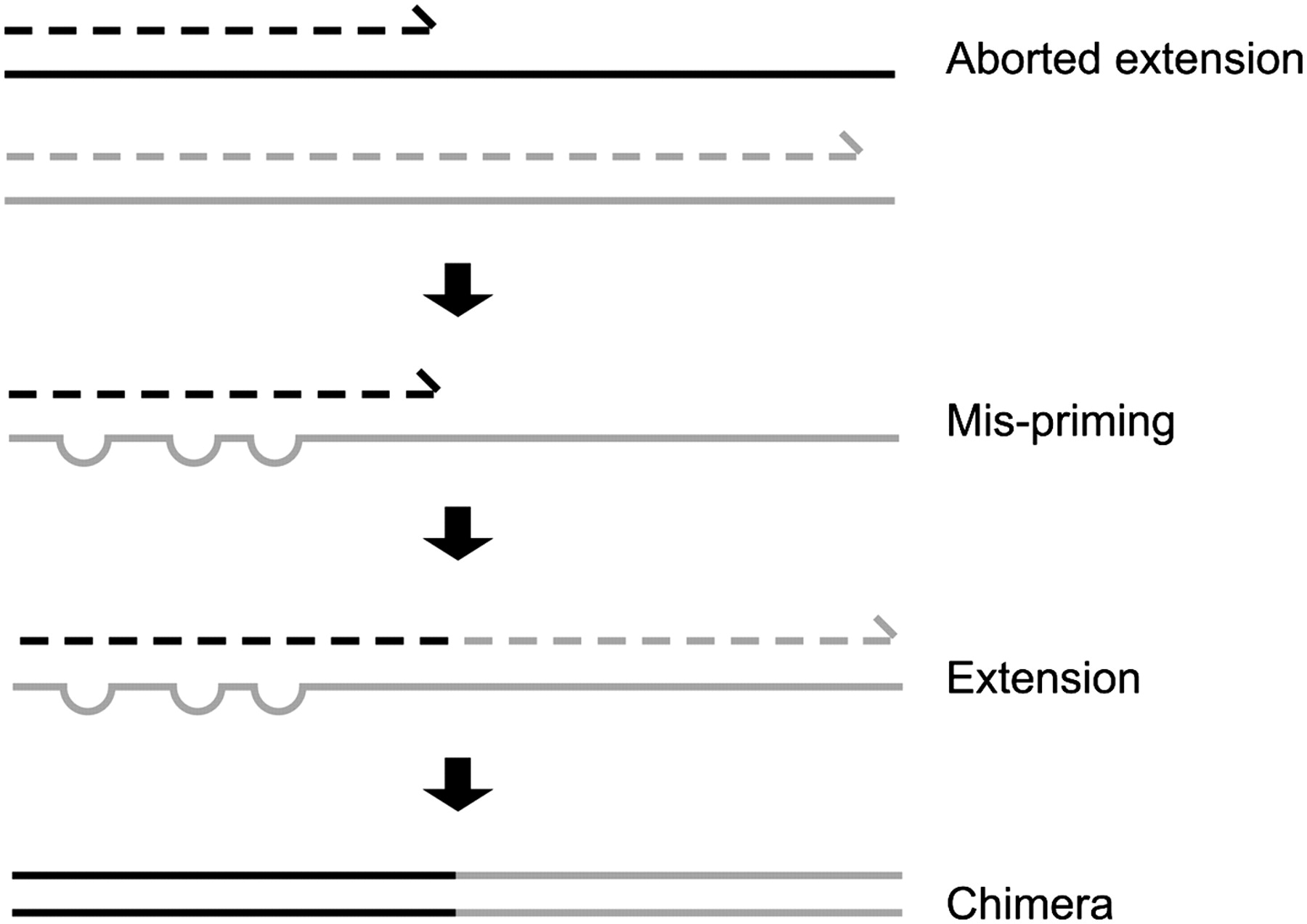
We will use the --uchime3_denovo algorithm as implemented in VSEARCH for removing chimeric sequences from our denoised reads.
nano chimera
#!/bin/bash
source moduleload
cd ../output/
vsearch --uchime3_denovo denoised.fasta --sizein --relabel zotu. --nonchimeras ../final/zotus.fasta
chmod +x chimera
./chimera
vsearch v2.21.1_linux_x86_64, 503.8GB RAM, 72 cores
https://github.com/torognes/vsearch
Reading file denoised.fasta 100%
9907 nt in 49 seqs, min 189, max 213, avg 202
Masking 100%
Sorting by abundance 100%
Counting k-mers 100%
Detecting chimeras 100%
Found 17 (34.7%) chimeras, 32 (65.3%) non-chimeras,
and 0 (0.0%) borderline sequences in 49 unique sequences.
Taking abundance information into account, this corresponds to
448 (0.4%) chimeras, 110965 (99.6%) non-chimeras,
and 0 (0.0%) borderline sequences in 111413 total sequences.
The output shows that 99.6% of our denoised reads were kept and 32 sequences were incorporated into our final ZOTU list.
The zotus.fasta file is the first output file we have created that we need for our statistical analysis!
5. Count table
Now that we have created our list of biologically relevant sequences or ZOTUs or “species”, we are ready to generate a count table, also known as a frequency table. This will be the last step in our bioinformatic analysis pipeline. A count table is something you might have encountered before during some traditional ecological surveys you have conducted, whereby a table was created with site names as column headers, a species list as rows, and the number in each cell representing the number of individuals observed for a specific species at a specific site.
In metabarcoding studies, a count table is analogous, where it tells us how many times each sequence has appeared in each sample. It is the end-product of all the bioinformatic processing steps we have conducted today. Now that we have identified what we believe to be true biological sequences, we are going to generate our count table by matching the merged sequences to our ZOTU sequence list. Remember that the sequences within the filtered.fasta file have the information of the sample they belong to present in the sequence header, which is how the --usearch_global command in VSEARCH can generate the count table. The --db parameter allows us to set the ZOTU sequence list (zotus.fasta) as the database to search against, while we can specify the --strand parameter as plus, since all sequences are in the same direction after primer trimming. Finally, we need to incorporate a 97% identity threshold for this function through the --id parameter. This might seem counter-intuitive, since we employed a denoising approach. However, providing some leniency on which sequences can map to our ZOTU will allow us to incorporate a larger percentage of the data set. As some ZOTUs might be more similar to each other than 97%, the algorithm will sort out the best match and add the sequence to the correct ZOTU sequence. If you’d like to be more conservative, you can set this threshold to 99%, though this is not recommended by the authors.
nano table
#!/bin/bash
source moduleload
cd ../output/
vsearch --usearch_global filtered.fasta --db ../final/zotus.fasta --strand plus --id 0.97 --otutabout ../final/zotutable.txt
chmod +x table
./table
vsearch v2.21.1_linux_x86_64, 503.8GB RAM, 72 cores
https://github.com/torognes/vsearch
Reading file ../final/zotus.fasta 100%
6476 nt in 32 seqs, min 189, max 213, avg 202
Masking 100%
Counting k-mers 100%
Creating k-mer index 100%
Searching 100%
Matching unique query sequences: 122120 of 123537 (98.85%)
Writing OTU table (classic) 100%
The output printed to the Terminal window shows that we managed to map 122,120 sequences to the ZOTU list, which were incorporated into our count table.
That’s it for the bioinformatic processing of our sequencing data, we can now move on to assigning a taxonomic ID to our ZOTUs!
Key Points
Quality filtering on sequence length, ambiguous basecalls, and maximum error rate
Clustering sequences into OTUs can miss closely related species
Denoising sequences can create overestimate diversity when reference databases are incomplete
Taxonomy assignment
Overview
Teaching: 30 min
Exercises: 60 minQuestions
What reference databases are available for metabarcoding
How do I make a custom database?
What are the main strategies for assigning taxonomy to OTUs?
What is the difference between Sintax and Naive Bayes?
Objectives
Create your own local reference database
Assign taxonomy using SINTAX
Assign taxonomy using local BLAST
Use
lessto explore a long help outputWrite a script to assign taxonomy
Taxonomy assignment
1. Introduction
As we have previously discussed, eDNA metabarcoding is the process whereby we amplify a specific gene region of the taxonomic group of interest from a DNA extract obtained from an environmental sample. Once the DNA is amplified, a library is prepared for sequencing. Up to this point, we have prepared the data for the bioinformatic process and executed a bioinformatic pipeline, which ended with a count table (zotutable.txt) and a ZOTU list (zotus.fasta) containing all the biologically relevant sequences. The next step in the bioinformatic process will be to assign a taxonomic ID to each sequence to determine what species were detected through our eDNA metabarcoding analysis.
There are four basic strategies to taxonomy classification based on (Hleap et al. (2021)) (and endless variations of each of these):
- Sequence Similarity (SS)
- Sequence Composition (SC)
- Phylogenetic (Ph)
- Probabilistic (Pr)
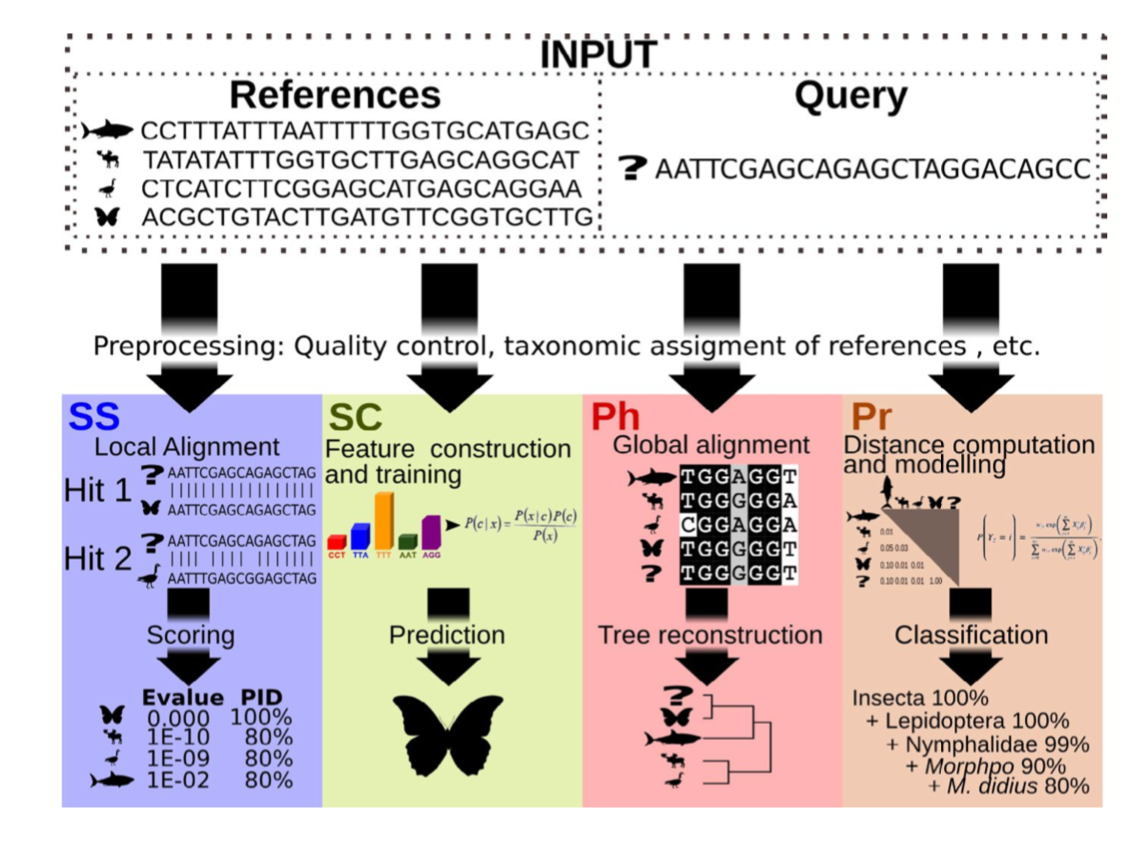
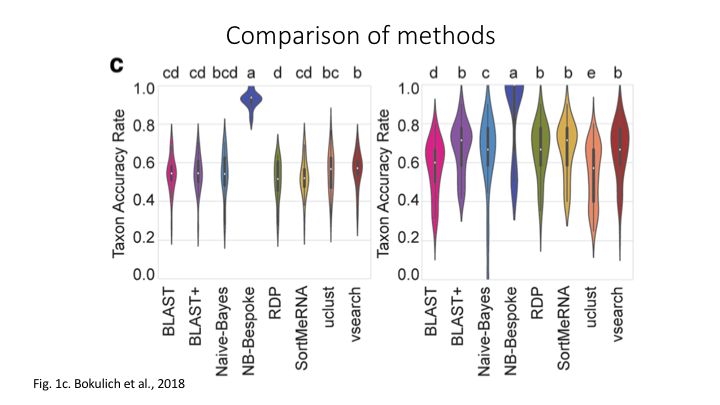
Of the four abovementioned strategies to taxonomy classification, the Sequence Similarity and Sequence Composition methods are currently the most frequently used for taxonomy assignment in the metabarcoding research community. Multiple comparative experiments have been conducted to determine the most optimal approach to assign a taxonomic ID to metabarcoding data, though it seems to be that the main conclusion of those papers is that their own method is best. One example of a comparative study is the one from Bokulich et al., 2018.
While there are multiple ways to assign a taxonomic ID to a sequence, all methods require a reference database to match your ZOTUs to a set of sequences with known taxonomic IDs. So, before we start with taxonomy assignment, let’s create our own local curated reference database using (CRABS), which stands for Creating Reference databases for Amplicon-Based Sequencing.
2. Reference databases
Several reference databases are available online. The most notable ones being NCBI, EMBL, BOLD, SILVA, and RDP. However, recent research has indicated a need to use custom curated reference databases to increase the accuracy of taxonomy assignment. While certain gene-specific or primer-specific reference databases are available (RDP: 16S microbial; MIDORI: COI eukaryotes; etc.), this essential data source is missing in most instances. We will, therefore, show you how to build your own custom curated reference database using CRABS.
For time-saving purposes, we will only build a small reference database that includes sharks (Chondrichthyes) for the primer set used during this experiment, rather than the reference database we will be using for taxonomy assignment later on that includes bony fish and sharks. However, the code provided here can be changed to build a reference database for any primer set or taxonomic group by changing some of the parameters. The final reference database that we will be using later on for taxonomy assignment is already placed in the refdb folder on NeSI.
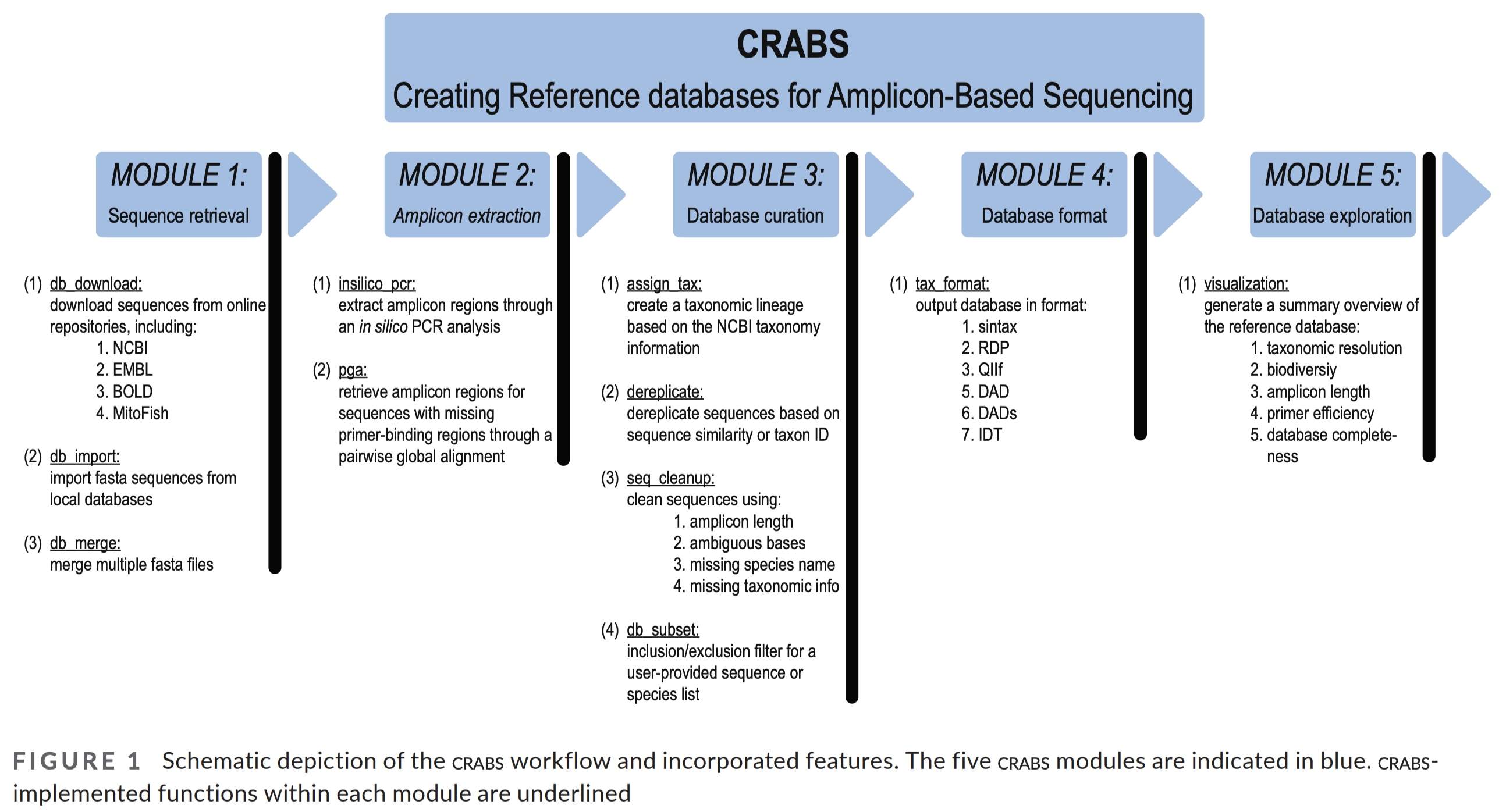
2.1 CRABS workflow
The CRABS workflow consists of seven parts, including:
- (1) download data from online repositories;
- (2) import downloaded data into CRABS format;
- (3) extract amplicons through in silico PCR analysis;
- (4) retrieving amplicons without primer-binding regions through pairwise global alignments;
- (5) curate and subset the local database;
- (6) export the local database in various taxonomic classifier formats; and
- (7) basic visualizations to explore the local reference database.
2.2 Help documentation
To bring up the help documentation for CRABS, we will first have to source the moduleload script to load the program.
source moduleload
crabs -h

2.3 Step 1: download data from online repositories
As a first step, we will download the 16S shark sequences from the NCBI online repository. While there are multiple repositories supported by CRABS, we’ll only focus on this one today, as NCBI is the most-widely used online repository in metabarcoding research. To download data from the NCBI servers, we need to provide a few parameters to CRABS. Maybe one of the trickiest to understand is the --query parameter. However, we can get the input string for this parameter from the NCBI website, as shown on the screenshot below.
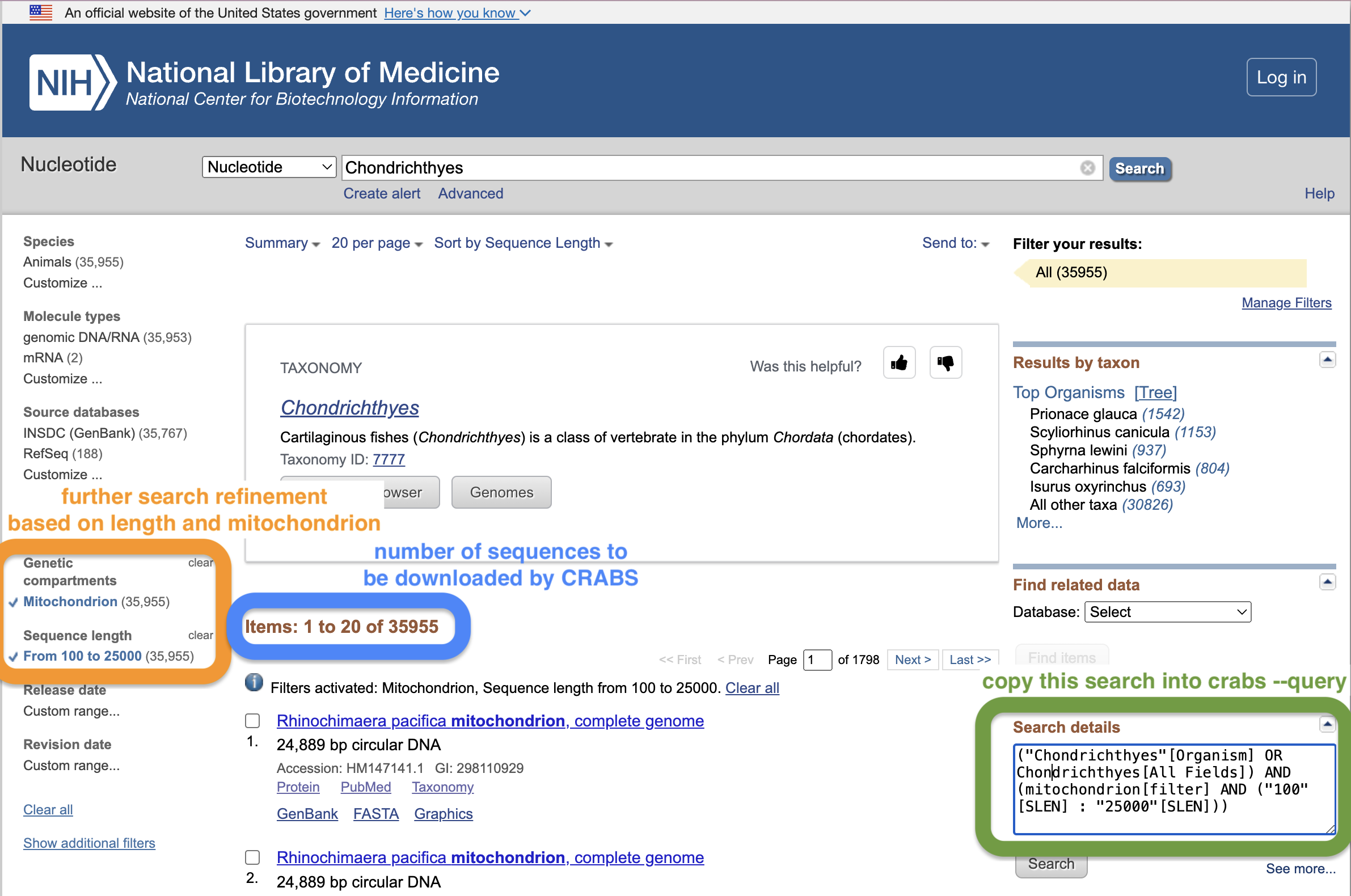
cd ../refdb/
crabs --download-ncbi --query '(16S[All Fields] AND ("Chondrichthyes"[Organism] OR Chondrichthyes[All Fields])) AND mitochondrion[filter]' --database nucleotide --email gjeunen@gmail.com --output ncbi_16S_chondrichthyes.fasta
/// CRABS | v1.0.6
| Function | Download NCBI database
|Retrieving NCBI info | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:01
| Downloading | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:23
| Results | Number of sequences downloaded: 3764/3764 (100.0%)
Let’s look at the top 10 lines of the downloaded fasta file to see what CRABS created:
head -n 10 ncbi_16S_chondrichthyes.fasta
>PP329120.1 Mobula hypostoma isolate MHBRsp_Gua mitochondrion, complete genome
GCTAGTGTAGCTTAATTTAAAGCATGGCACTGAAGATGCTAAGATAAAAATTAACCTTTTTCACAAGCAT
GAAGGTTTGGTCCCAGCCTCAATATTAGTTTTAACTTGACTTACACATGCAAGTCTCAGCATTCCGGTGA
GAACGCCCTAATCACCCCTAAAATTTGATTAGGAGCTGGTATCAGGCACATTCCAAGAACAGCCCATGAC
ACCTCGCTCAGCCACACCCACAAGGGAACTCAGCAGTGATAGATATTGTTCTATAAGCGCAAGCTTGAGA
CAATCAAAGTTATAAAGAGTTGGTTAATCTCGTGCCAGCCACCGCGGTTATACGAGTGACACAAATTAAT
ATTTCACGGCGTTAAGGGTGATTAGAAATAATCTCATCAGTATAAAGTTAAGACCTCATTAAGCTGTCAT
ACGCACTTATGATCGAAAACATCATTCACGAAAGTAACTTTACACAAACAGAATTTTTGACCTCACGACA
GTTAAGGCCCAAACTAGGATTAGATACCCTACTATGCTTAACCGCAAACATTGTTATTAATTAATACTAC
CTTAATACCCGCCTGAGTACTACAAGCGCTAGCTTAAAACCCAAAGGACTTGGCGGTGCTCCAAACCCAC
Here, we can see that the CRABS has downloaded the original format from the NCBI servers, which is a multi-line fasta document.
2.4 Step 2: import downloaded data into CRABS format
Since CRABS can download data from multiple online repositories, each with their own format, we need to first reformat the data to CRABS format before we can continue building our reference database. This reformatting can be done using the --import function, which will create a single line text document for each sequence, that contains a unique ID, the taxonomic lineage for that sequence, plus the sequence itself. The CRABS taxonomic lineage is based on the NCBI taxonomy database and requires 3 files to be downloaded using the --download-taxonomy function. However, to speed things up, these 3 files have already been provided to you.
crabs --import --import-format ncbi --names names.dmp --nodes nodes.dmp --acc2tax nucl_gb.accession2taxid --input ncbi_16S_chondrichthyes.fasta --output ncbi_16S_chondrichthyes.txt --ranks 'superkingdom;phylum;class;order;family;genus;species'
/// CRABS | v1.0.6
| Function | Import sequence data into CRABS format
| Read data to memory | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:05:29
|Phylogenetic lineage | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:00
|Fill missing lineage | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:00
| Results | Imported 3764 out of 3764 sequences into CRABS format (100.0%)
When we now look at the first line of the imported document, we can see how the structure has changed:
head -n 1 ncbi_16S_chondrichthyes.txt
PP329120 Mobula hypostoma 723540 Eukaryota Chordata Chondrichthyes Myliobatiformes Myliobatidae Mobula Mobula hypostoma GCTAGTGTAGCTTAATTTAAAGCATGGCACTGAAGATGCTAAGATAAAAATTAACCTTTTTCACAAGCATGAAGGTTT...
2.5 Step 3: extract amplicons through in silico PCR
Once we have formatted the data, we can start with a first curation step of the reference database, i.e., extracting the amplicon sequences. We will be extracting the amplicon region from each sequence through an in silico PCR analysis. With this analysis, we will locate the forward and reverse primer binding regions and extract the sequence in between. This will significantly reduce file sizes when larger data sets are downloaded, while also ensuring that only the necessary information is kept.
crabs --in-silico-pcr --input ncbi_16S_chondrichthyes.txt --output ncbi_16S_chondrichthyes_amplicon.txt --forward GACCCTATGGAGCTTTAGAC --reverse CGCTGTTATCCCTADRGTAACT
/// CRABS | v1.0.6
| Function | Extract amplicons through in silico PCR
| Import data | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:00
| Transform to fasta | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:00
| In silico PCR | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:00
| Exporting data | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:00
| Results | Extracted 3427 amplicons from 3764 sequences (91.05%)
2.6 Step 4: extract amplicons through pairwise global alignments
Amplicons in the originally downloaded sequences might be missed during the in silico PCR analysis when one or both primer-binding regions are not incorporated in the online deposited sequence. This can happen when the reference barcode is generated with the same primer set or if the deposited sequence is incomplete in the primer-binding regions (denoted as “N” in the sequence). To retrieve those missed amplicons, we can use the already-retrieved amplicons as seed sequences in a Pairwise Global Alignment (PGA) analysis.
crabs --pairwise-global-alignment --input ncbi_16S_chondrichthyes.txt --amplicons ncbi_16S_chondrichthyes_amplicon.txt --output ncbi_16S_chondrichthyes_pga.txt --forward GACCCTATGGAGCTTTAGAC --reverse CGCTGTTATCCCTADRGTAACT --percent-identity 0.9 --coverage 90
/// CRABS | v1.0.6
| Function | Retrieve amplicons without primer-binding regions
| Import data | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:00
| Transform to fasta | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:00
| Pairwise alignment | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 0% -:--:-- 0:00:01
|Parse alignment data | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:00
| Exporting data | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:00
| Results | Retrieved 42 amplicons without primer-binding regions from 337 sequences
2.7 Step 5: dereplicating the database
Now that we have retrieved all amplicons from the downloaded sequences, we can remove duplicate sequences for each species, i.e., dereplicating the reference database. This step helps reduce the file size even more, plus is essential for most taxonomic classifiers that restrict the output to N number of best hits.
crabs --dereplicate --input ncbi_16S_chondrichthyes_pga.txt --output ncbi_16S_chondrichthyes_derep.txt --dereplication-method unique_species
/// CRABS | v1.0.6
| Function | Dereplicate CRABS database
| Import data | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:00
| Exporting data | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:00
| Results | Written 723 unique sequences to ncbi_16S_chondrichthyes_derep.txt out of 3469 initial sequences (20.84%)
2.8 Step 6: filter the reference database
The final curation step for our custom reference database is to clean up the database using a variety of parameters, including minimum length, maximum length, maximum number of ambiguous base calls, environmental sequences, sequences for which the species name is not provided, and sequences with unspecified taxonomic levels.
crabs --filter --input ncbi_16S_chondrichthyes_derep.txt --output ncbi_16S_chondrichthyes_clean.txt --minimum-length 150 --maximum-length 250 --environmental --no-species-id --maximum-n 1 --rank-na 2
/// CRABS | v1.0.6
| Function | Filter CRABS database
| Included parameters | "--minimum-length", "--maximum-length", "--maximum-n", "--environmental", "--no-species-id", "--rank-na"
| Import data | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:00
| Exporting data | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:00
| Results | Written 706 filtered sequences to ncbi_16S_chondrichthyes_clean.txt out of 723 initial sequences (97.65%)
| | Maximum ambiguous bases filter: 17 sequences not passing filter (2.35%)
2.9 Step 7: export the reference database to a format supported by the taxonomic classifier software
Now that we have created a custom curated reference database containing 706 shark sequences for the 16S rRNA gene, we can export the reference database to be used by different taxonomic classifiers. Multiple formats are implemented in CRABS, but today we will be exporting the reference database to SINTAX format, which is a kmer based classifier implemented in USEARCH and VSEARCH.
crabs --export --input ncbi_16S_chondrichthyes_clean.txt --output ncbi_16S_chondrichthyes_sintax.fasta --export-format sintax
/// CRABS | v1.0.6
| Function | Export CRABS database to SINTAX format
| Import data | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:00
| Exporting data | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:00
| Results | Written 706 sequences to ncbi_16S_chondrichthyes_sintax.fasta out of 706 initial sequences (100.0%)
Let’s open the reference database to see what the final database looks like!
head -n 6 ncbi_16S_chondrichthyes_sintax.fasta
>PP329120;tax=d:Eukaryota,p:Chordata,c:Chondrichthyes,o:Myliobatiformes,f:Myliobatidae,g:Mobula,s:Mobula_hypostoma
ACTTAAGTCACTTTTAAGATATTAAATTTTTTCCTCAGGGTATAAAATAACAAACTAATTTTCTGACTTAACCTTGTTTTTGGTTGGGGCGACCGAGGGGTAAAACAAAACCCCCTTATCGAATGTGTGAAATATCACTTAAAAATTAGAGTTACATCTCTAGTTAATAGAAAATCTAACGAACAATGACCCAGGAAACTTATCCTGATCAATGAACCA
>PQ469261;tax=d:Eukaryota,p:Chordata,c:Chondrichthyes,o:Carcharhiniformes,f:Carcharhinidae,g:Prionace,s:Prionace_glauca
ACTTAAATTAATTATGTAACCATCCATTTCCCAGGAAATAAACAAAATATACAACACTTCTAATTTAACTGTTTTTGGTTGGGGTGACCAAGGGGAAAAACAAATCCCCCTCATCGATTGAGTACTAAGTACTTAAAAATTAGAATGACAATTCTAATTAATAAAACATTTACCGAAAAATGATCCAGGATTTCCTGATCAATGAACCA
>PQ182775;tax=d:Eukaryota,p:Chordata,c:Chondrichthyes,o:Carcharhiniformes,f:Triakidae,g:Mustelus,s:Mustelus_schmitti
ACTTAAGTTAATTATGTAACCATTTATTCCTTAGGGTATAAACAAAATATACAATACTTCTAACTTAACTGTTTTTGGTTGGGGTGACCGAGGGGGAAAACAAATCCCCCTCATCGATTGAGTACTCAGTACTTGAAAATCAGAATAACAATTCTGATTAATAAAATATTTAGCGAAAAATGACCCAGGATTTCCTGATCAATGAACCA
That’s all it takes to generate your own reference database. We are now ready to assign a taxonomy to our OTUs that were generated in the previous section!
3. The 16S Vertebrate reference database
For our tutorial data set, we will be using the sintax_vertebrata_16S.fasta and vertebrata_16S databases, with the first one being in SINTAX format and the second one being in BLAST format. The reference databases were created using CRABS, as shown above, and contain data from the MitoFish online repository, NCBI online repository, and local New Zealand barcodes that we generated at the University of Otago. These reference databases hold 57,092 high quality, unique, 16S rRNA reference barcodes for vertebrate taxa.
4. Taxonomic classification
4.1 SINTAX
The first method we will explore is the SINTAX algorithm. The SINTAX algorithm predicts taxonomy by using k-mer similarity to identify the top hit in a reference database and provides bootstrap confidence for all ranks in the prediction. Unlike the RDP classifier, SINTAX does not rely on Bayesian posteriors. Hence, training the dat aset is not required and results can be obtained very quickly. SINTAX was developed for USEARCH and is also incorporated into VSEARCH through the --sintax command. More information about the SINTAX algorithm can be found on the USEARCH website. When thinking about the four basic strategies for taxonomy assignment, SINTAX belongs to the Sequence Composition (SC) category.
cd ../scripts/
nano sintax
#!/bin/bash
source moduleload
cd ../final/
vsearch --sintax zotus.fasta --db ../refdb/sintax_vertebrata_16S.fasta --sintax_cutoff 0.8 --tabbedout sintax_taxonomy.txt
chmod +x sintax
./sintax
vsearch v2.21.1_linux_x86_64, 503.8GB RAM, 72 cores
https://github.com/torognes/vsearch
Reading file ../refdb/sintax_vertebrata_16S.fasta 100%
WARNING: 1 invalid characters stripped from FASTA file: ?(1)
REMINDER: vsearch does not support amino acid sequences
10986499 nt in 57092 seqs, min 150, max 249, avg 192
Counting k-mers 100%
Creating k-mer index 100%
Classifying sequences 100%
Classified 26 of 32 sequences (81.25%)
Taxonomy assignment through SINTAX was able to classify 27 out of 32 OTUs. Let’s use the head command to inspect the output file.
head ../final/sintax_taxonomy.txt
zotu.3 d:Eukaryota(1.00),p:Chordata(1.00),c:Actinopteri(1.00),o:Perciformes(1.00),f:Bovichtidae(1.00),g:Bovichtus(1.00),s:Bovichtus_variegatus(1.00) + d:Eukaryota,p:Chordata,c:Actinopteri,o:Perciformes,f:Bovichtidae,g:Bovichtus,s:Bovichtus_variegatus
zotu.1 d:Eukaryota(1.00),p:Chordata(1.00),c:Actinopteri(1.00),o:Scombriformes(1.00),f:Gempylidae(1.00),g:Thyrsites(1.00),s:Thyrsites_atun(1.00) + d:Eukaryota,p:Chordata,c:Actinopteri,o:Scombriformes,f:Gempylidae,g:Thyrsites,s:Thyrsites_atun
zotu.5 d:Eukaryota(1.00),p:Chordata(1.00),c:Actinopteri(1.00),o:Labriformes(1.00),f:Labridae(1.00),g:Notolabrus(0.89),s:Notolabrus_fucicola(0.86) + d:Eukaryota,p:Chordata,c:Actinopteri,o:Labriformes,f:Labridae,g:Notolabrus,s:Notolabrus_fucicola
zotu.4 d:Eukaryota(1.00),p:Chordata(1.00),c:Actinopteri(1.00),o:Blenniiformes(1.00),f:Tripterygiidae(1.00),g:Forsterygion(1.00),s:Forsterygion_lapillum(0.99) + d:Eukaryota,p:Chordata,c:Actinopteri,o:Blenniiformes,f:Tripterygiidae,g:Forsterygion,s:Forsterygion_lapillum
zotu.6 d:Eukaryota(1.00),p:Chordata(1.00),c:Actinopteri(1.00),o:Blenniiformes(1.00),f:Tripterygiidae(1.00),g:Forsterygion(1.00),s:Forsterygion_lapillum(0.91) + d:Eukaryota,p:Chordata,c:Actinopteri,o:Blenniiformes,f:Tripterygiidae,g:Forsterygion,s:Forsterygion_lapillum
zotu.2 d:Eukaryota(1.00),p:Chordata(1.00),c:Actinopteri(1.00),o:Mugiliformes(1.00),f:Mugilidae(1.00),g:Aldrichetta(1.00),s:Aldrichetta_forsteri(1.00) + d:Eukaryota,p:Chordata,c:Actinopteri,o:Mugiliformes,f:Mugilidae,g:Aldrichetta,s:Aldrichetta_forsteri
zotu.12 d:Eukaryota(1.00),p:Chordata(1.00),c:Actinopteri(1.00),o:Myctophiformes(1.00),f:Myctophidae(1.00),g:Hygophum(1.00),s:Hygophum_hanseni(1.00) + d:Eukaryota,p:Chordata,c:Actinopteri,o:Myctophiformes,f:Myctophidae,g:Hygophum,s:Hygophum_hanseni
zotu.11 d:Eukaryota(1.00),p:Chordata(1.00),c:Actinopteri(1.00),o:Osmeriformes(1.00),f:Retropinnidae(1.00),g:Retropinna(1.00),s:Retropinna_retropinna(1.00) + d:Eukaryota,p:Chordata,c:Actinopteri,o:Osmeriformes,f:Retropinnidae,g:Retropinna,s:Retropinna_retropinna
zotu.9
zotu.13 d:Eukaryota(1.00),p:Chordata(1.00),c:Actinopteri(1.00),o:Scombriformes(1.00),f:Centrolophidae(1.00),g:Seriolella(0.99),s:Seriolella_brama(0.99) + d:Eukaryota,p:Chordata,c:Actinopteri,o:Scombriformes,f:Centrolophidae,g:Seriolella,s:Seriolella_brama
4.2 BLAST
The second and last method we will explore is BLAST (Basic Local Alignment Search Tool), probably the most well known and most widely used method to assign a taxonomic ID to sequences. As the name suggests, BLAST assigns a taxonomic ID to a sequence through best hits from local alignments against a reference database. Hence, when thinking about the four basic strategies for taxonomy assignment, BLAST belongs to the Sequence Similarity (SS) category. While BLAST can be run online and remotely against the full NCBI database, BLAST can also be run locally on a curated reference database. Since CRABS can download data from multiple online repositories, running BLAST locally will aid in increased taxonomic assignment accuracy by incorporating a larger number of barcodes.
nano blast
#!/bin/bash
source moduleload
cd ../refdb/
blastn -query ../final/zotus.fasta -db vertebrata_16S -max_target_seqs 1 -perc_identity 50 -qcov_hsp_perc 50 -outfmt "6 qaccver saccver staxid sscinames length pident mismatch qcovs evalue bitscore qstart qend sstart send gapopen" -out ../final/blast_taxonomy.txt
chmod +x blast
./blast
Running BLAST locally doesn’t produce any text in the Terminal Window, but we can use the head command to inspect the output file.
head -n 32 ../final/blast_taxonomy.txt
zotu.1 OP168897 499858 Thyrsites atun 205 100.000 0 100 9.00e-106 379 1 205 1 205 0
zotu.2 JQ060654 443748 Aldrichetta forsteri 202 100.000 0 100 4.12e-104 374 1 202 1 202 0
zotu.3 MF991327 36205 Bovichtus variegatus 203 100.000 0 100 1.15e-104 375 1 203 1 203 0
zotu.4 AY141416 206115 Forsterygion lapillum 203 98.030 4 100 5.40e-98 353 1 203 1 203 0
zotu.5 EU848435 548164 Notolabrus fucicola 205 99.512 1 100 4.19e-104 374 1 205 1 205 0
zotu.6 AY141416 206115 Forsterygion lapillum 203 99.507 1 100 5.36e-103 370 1 203 1 203 0
zotu.7 AY520117 8210 Notothenia angustata 202 100.000 0 100 4.12e-104 374 1 202 1 202 0
zotu.8 NC_016669 691800 Sprattus muelleri 202 100.000 0 100 4.12e-104 374 1 202 1 202 0
zotu.9 MK335855 1311454 Istigobius ornatus 159 87.421 18 80 2.62e-46 182 40 197 39 196 2
zotu.10 GQ365289 159454 Hyporhamphus melanochir 203 100.000 0 100 1.15e-104 375 1 203 1 203 0
zotu.11 MZ301925 170203 Retropinna retropinna 205 100.000 0 100 9.00e-106 379 1 205 1 205 0
zotu.12 AB024914 89989 Hygophum hanseni 202 100.000 0 100 4.12e-104 374 1 202 1 202 0
zotu.13 AB205416 316134 Seriolella caerulea 205 100.000 0 100 9.00e-106 379 1 205 1 205 0
zotu.14 EU848442 443792 Lotella rhacina 200 96.500 7 100 2.49e-91 331 1 200 1 200 0
zotu.15 NC_080276 2972643 Salmo pallaryi 205 100.000 0 100 9.00e-106 379 1 205 1 205 0
zotu.16 KP411798 879484 Enneanectes reticulatus 149 85.906 20 73 4.57e-39 158 56 203 47 195 1
zotu.17 AY141416 206115 Forsterygion lapillum 205 85.854 27 100 7.47e-57 217 1 205 1 203 1
zotu.18 AY141416 206115 Forsterygion lapillum 203 97.537 5 100 2.51e-96 348 1 203 1 203 0
zotu.19 LC627613 107245 Lepidopus caudatus 204 100.000 0 99 3.24e-105 377 1 204 1 204 0
zotu.20 AY141416 206115 Forsterygion lapillum 205 86.341 26 100 1.61e-58 222 1 205 1 203 1
zotu.21 NC_016673 691801 Sprattus antipodum 202 99.505 1 100 1.92e-102 368 1 202 1 202 0
zotu.22 AY279764 241364 Odax pullus 201 100.000 0 100 1.47e-103 372 1 201 1 201 0
zotu.23 PP661061 2791166 Pseudophycis palmata 198 91.919 16 100 3.25e-75 278 1 198 1 198 0
zotu.24 AY141416 206115 Forsterygion lapillum 205 95.610 7 100 3.31e-90 327 1 205 1 203 1
zotu.25 NC_088030 1241940 Umbrina canosai 154 84.416 12 71 4.83e-34 141 62 213 59 202 6
zotu.26 KY020202 879906 Xiphasia setifer 132 78.030 26 68 9.44e-16 80.5 56 184 66 197 3
zotu.27 OM470922 2740763 Lepadogaster candolii 147 83.673 21 72 2.11e-32 135 56 200 54 199 3
zotu.28 PP661061 2791166 Pseudophycis palmata 198 91.414 17 100 1.51e-73 272 1 198 1 198 0
zotu.29 NC_016669 691800 Sprattus muelleri 202 98.515 3 100 4.15e-99 357 1 202 1 202 0
zotu.30 KY020202 879906 Xiphasia setifer 132 78.030 26 68 9.44e-16 80.5 56 184 66 197 3
zotu.31 NC_037961 2605477 Dipturus nasutus 210 100.000 0 100 1.54e-108 388 1 210 1 210 0
zotu.32 CRABS:Acanthoclinus_fuscus 2696578 Acanthoclinus fuscus 203 97.537 4 100 2.51e-96 348 1 203 1 202 1
Using the head command shows us that BLAST managed to provide a taxonomic ID to all 32 ZOTU sequences in our data, unlike SINTAX, which managed to provide a taxonomic ID to only 27 ZOTUs. However, if we look at the output, the BLAST results are “low” in similarity and coverage for the ZOTUs for which SINTAX does not provide any taxonomic ID.
Please note that I do not recommend working on solely top BLAST hits, as we did in our example. This choice was done as a time-saving approach. I recommend setting the parameter --max-target-seqs to 50 or 100 and calculate the Most Recent Common Ancestor (MRCA) based on those BLAST results. While this is outside the scope of this workshop, there are some useful programs that are publicly available that can do this for you, e.g., ALEX (Ancestor Link EXplorer).
Key Points
The main taxonomy assignment strategies are global alignments, local alignments, and machine learning approaches
Local curated reference databases increase taxonomic assignment accuracy
Data curation
Overview
Teaching: 15 min
Exercises: 30 minQuestions
What type of artefacts are still present in the data?
How to process ZOTUs present in negative control samples?
How to test if data needs to be rarefied?
Objectives
Remove artefacts using tombRaider
Plot rarefaction curves and calculate rarefaction indices
Data curation
1. Introduction
At this point, we have created a file containing a list of ZOTUs (zotus.fasta), a count table (zotutable.txt), and a taxonomic ID for each ZOTU (blast_taxonomy.txt). Before exploring the data and conducting the statistical analysis, we need to perform some data curation steps to further clean the data. Within the eDNA metabarcoding research community, there is not yet a standardised manner of data curation. However, steps that are frequently included are (list is not in order of importance):
- Abundance filtering of specific detections to eliminate the risk of barcode hopping signals interfering with the analysis.
- Either involves removing singleton detections or applying an abundance threshold.
- Filtering of ZOTUs which have a positive detection in the negative control samples, either by:
- Complete removal of ZOTUs.
- Applying an abundance threshold obtained from the read count in the negative controls.
- A statistical test.
- Removal of low-confidence taxonomy assignments (if analysis is taxonomy-dependent).
- Discarding samples with low read count.
- Removal of artefacts.
- Transformation of raw read count to relative abundance, presence-absence, or other transformations.
2. Artefact removal through tombRaider
As a first data curation step, we will be removing spurious artefacts using a novel algorithm developed by our team, i.e., tombRaider. Although stringent quality filtering parameters have been employed during the bioinformatic processing steps, artefacts still persist and are known to inflate diversity estimates. These artefacts might arise from the incorporation of PCR errors or the presence of pseudogenes. For more information, please consult the tombRaider manuscript.
tombRaider can identify and remove artefacts using a single line of code, based on taxonomic ID, sequence similarity, the presence of stop codons, and co-occurrence patterns.
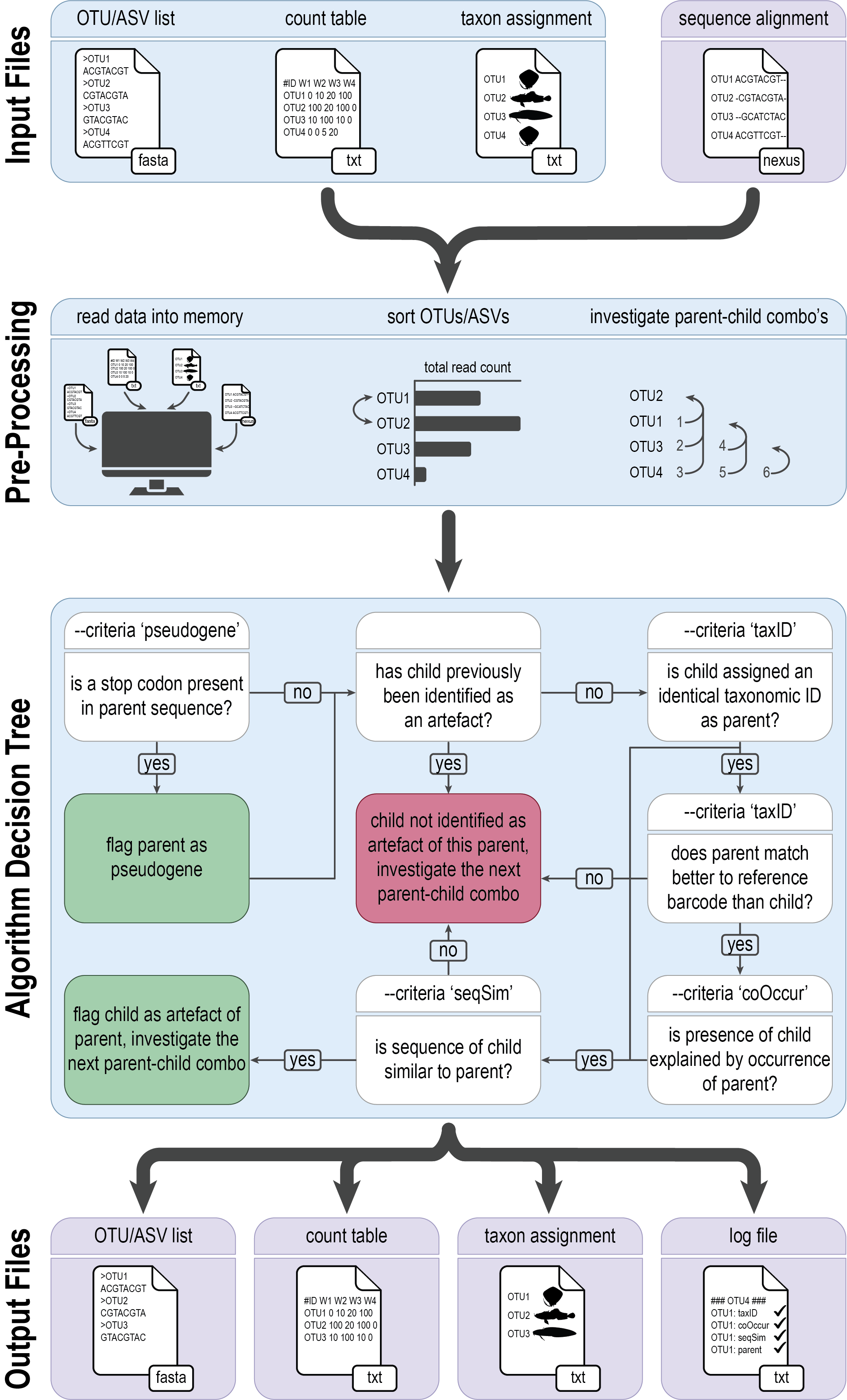
To execute tombRaider, we need to provide the count table, list of ZOTUs, and taxonomy file as input files, while also specifying the novel output files where artefacts are removed.
nano tombraider
#!/bin/bash
source moduleload
cd ../final/
tombRaider --criteria 'taxId;seqSim;coOccur' --frequency-input zotutable.txt --sequence-input zotus.fasta --taxonomy-input blast_taxonomy.txt --blast-format '6 qaccver saccver staxid sscinames length pident mismatch qcovs evalue bitscore qstart qend sstart send gapopen' --frequency-output zotutable_new.txt --sequence-output zotus_new.fasta --taxonomy-output blast_taxonomy_new.txt --log tombRaider_log.txt --occurrence-type abundance --occurrence-ratio 'count;1' --sort 'total read count' --similarity 90
chmod +x tombraider
./tombraider
/// tombRaider | v1.0
| Included Criteria | taxid, seqsim, cooccur
| Excluded Criteria | pseudogene
| Reading Files | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:00
| Identify artefacts | ━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━━ 100% 0:00:00 0:00:01
| Summary Statistics |
| Total # of ASVs | 32
|Total # of Artefacts | 6 (18.75%)
| Artefact List |
| parent: zotu.4 | children: zotu.18, zotu.24
| parent: zotu.8 | child: zotu.29
| parent: zotu.17 | child: zotu.20
| parent: zotu.23 | child: zotu.28
| parent: zotu.26 | child: zotu.30
From the output we can see that tombRaider has identified 6 artefacts, which have now been merged with their parent sequence.
3. Contamination removal with R
The next step of data curation is to investigate what is observed in our negative control samples and remove any potential contamination in our data.
In our tutorial data, all reads assigned to our negative control sample dropped out after quality filtering. Hence, we wouldn’t be conducting this step for our data normally. However, to show you how to remove contamination, we can generate mock data that resembles a negative control sample.
Please note that it is at this point we will start using the R environment, rather than the Terminal Window.
As of yet, there is no consensus in the metabarcoding research community on how to properly remove contamination from a data set. In this workshop, I will make use of the R package microDecon to remove the contamination signal from the data. However, please also investigate the R package Decontam, which is associated with the popular R package phyloseq. Another option you can investigate is the use of an abundance threshold, whereby you calculate the highest row-wise relative abundance for a positive detection in the negative control samples and remove all positive detections that fall below this relative abundance threshold.
3.1 Setting up the R Environment
Open RStudio from the launcher
## set working directory
setwd("~/obss_2024/edna/final")
## add the workshop library
.libPaths(c(.libPaths(), "/nesi/project/nesi02659/obss_2024/R_lib"))
## load libraries
library(microDecon)
library(ggplot2)
library(dplyr)
library(car)
library(lattice)
library(dplyr)
library(ggExtra)
library(ampvis2)
library(dplyr)
library(DivE)
library(sf)
library(iNEXT.3D)
3.2 Decontamination
## read data into R
count_table <- read.table('zotutable_new.txt', header = TRUE, sep = '\t', check.names = FALSE, comment.char = '')
taxonomy_table <- read.table('blast_taxonomy_new.txt', header = FALSE, sep = '\t', check.names = FALSE, comment.char = '')
## add mock data representing a negative control sample
count_table$neg <- 0
count_table$neg[sample(1:nrow(count_table), 3)] <- sample(0:10, 3, replace = TRUE)
count_table <- count_table[, c(1, which(names(count_table) == 'neg'), setdiff(2:ncol(count_table), which(names(count_table) == 'neg')))]
## add taxonomy information to count_table
names(count_table)[names(count_table) == "#OTU ID"] <- "V1"
count_table <- merge(count_table, taxonomy_table[, c("V1", "V4")], by = "V1", all.x = TRUE)
# run microDecon
decontaminated <- decon(count_table, numb.blanks = 1, numb.ind = c(6,5), taxa = TRUE)
4. Rarefaction
The second to last data curation step we will introduce during this workshop is how to deal with differences in sequencing depth between samples and assess if sufficient sequencing depth was achieved for each sample. With the realisation that a higher read count could lead to a larger number of ZOTUs being detected, there has been much debate in the scientific community on how to deal with this issue. Some studies argue that it is essential to “level the playing field” and use the same number of sequences across all samples. This random subsampling of the data is called rarefaction. See Cameron et al., 2021 as an example. Other studies, on the other hand, argue vehemently agains such an approach. For example, you can read McMurdie et al., 2014 for more on this topic. While this debate is still ongoing, I suggest to run a couple of statistical tests and visualisations that can help determine the need to rarefy your data. Afterwards, you can make an informed decision and clearly state why or why not you have decided to process your data in a certain way.
I suggest to run 4 tests to determine if rarefying your data might be necessary, including (1) determine if there is a big difference in read count between samples, (2) determine if there is a positive correlation between sequencing depth and number of observed ZOTUs, (3) draw so-called rarefaction curves to determine if sufficient sequencing coverage has been achieved, and (4) calculate curvature indices to provide a statistical test regarding the observed patterns in rarefaction curves.
As stated above, all of the analyses will be conducted in R.
4.1 Read distribution
## read data into R
count_table <- read.table('zotutable_new.txt', header = TRUE, sep = '\t', row.names = 1, check.names = FALSE, comment.char = '')
metadata_table <- read.table('../stats/metadata.txt', header = TRUE, sep = '\t', row.names = 1, check.names = FALSE, comment.char = '')
## add column sums of count_table to metadata_table
metadata_table$total_read_count <- colSums(count_table)[rownames(metadata_table)]
## plot total read count in decreasing order
sample_colors <- c("mudflats" = "#EAE0DA", "shore" = "#A0C3D2")
ggplot(metadata_table, aes(x = reorder(row.names(metadata_table), -total_read_count), y = total_read_count, fill = Location)) +
geom_bar(stat = "identity") +
labs(x = "Location", y = "Total Read Count") +
theme_classic() +
scale_fill_manual(values = sample_colors) +
scale_y_log10(breaks = 10^(seq(0, 6, by = 2)), labels = scales::comma) +
theme(axis.text.x = element_text(angle = 90, hjust = 1), legend.position = "bottom")
ggplot(metadata_table, aes(x = reorder(row.names(metadata_table), -total_read_count), y = total_read_count, fill = Location)) +
geom_bar(stat = "identity") +
labs(x = "Location", y = "Total Read Count") +
theme_classic() +
scale_fill_manual(values = sample_colors) +
theme(axis.text.x = element_text(angle = 90, hjust = 1), legend.position = "bottom")
## print basic stats of read distribution across samples
cat("Maximum value:", max(metadata_table$total_read_count, na.rm = TRUE), "in sample:", rownames(metadata_table)[which.max(metadata_table$total_read_count)], "\n")
cat("Minimum value:", min(metadata_table$total_read_count, na.rm = TRUE), "in sample:", rownames(metadata_table)[which.min(metadata_table$total_read_count)], "\n")
cat("Mean value:", mean(metadata_table$total_read_count, na.rm = TRUE), "\n")
cat("Standard error:", sd(metadata_table$total_read_count, na.rm = TRUE) / sqrt(sum(!is.na(metadata_table$total_read_count))), "\n")
## print basic stats of read distribution between groups
metadata_table %>%
group_by(Location) %>%
summarise(
mean_count = mean(total_read_count, na.rm = TRUE),
se_count = sd(total_read_count, na.rm = TRUE) / sqrt(n())
)
## run t-test (Welch's t-test, given distribution differences), and plot data for visualisation
metadata_table$total_read_count[is.na(metadata_table$total_read_count)] <- 0
bartlett.test(total_read_count ~ Location, data = metadata_table)
histogram(~ total_read_count | Location, data = metadata_table, layout = c(1,2))
t.test(total_read_count ~ Location, data = metadata_table, var.equal = FALSE, conf.level = 0.95)
ggplot(metadata_table, aes(x = Location, y = total_read_count, fill = Location)) +
geom_boxplot() +
labs(x = "Location", y = "Total Read Count") +
theme_classic() +
scale_fill_manual(values = sample_colors) +
theme(axis.text.x = element_text(angle = 45, hjust = 1))
4.2 Sequencing depth and detected ZOTUs correlation
## read data into R
count_table <- read.table('zotutable_new.txt', header = TRUE, sep = '\t', row.names = 1, check.names = FALSE, comment.char = '')
taxonomy_table <- read.table('blast_taxonomy_new.txt', header = TRUE, sep = '\t', row.names = 1, check.names = FALSE, comment.char = '')
metadata_table <- read.table('../stats/metadata.txt', header = TRUE, sep = '\t', row.names = 1, check.names = FALSE, comment.char = '')
## add column sums of count_table to metadata_table
metadata_table$total_read_count <- colSums(count_table)[rownames(metadata_table)]
metadata_table$total_observations <- colSums(count_table > 0)[rownames(metadata_table)]
# fit the GLM model
glm_model <- glm(total_observations ~ total_read_count,
family = poisson(link = "log"), data = metadata_table)
summary(glm_model)
# make predictions
metadata_table$predicted <- predict(glm_model, type = "response")
# plot results
metadata_table$Location <- as.factor(metadata_table$Location)
sample_colors <- c("mudflats" = "#EAE0DA", "shore" = "#A0C3D2")
p <- ggplot(metadata_table, aes(x = total_read_count, y = total_observations, color = Location)) +
geom_point(aes(shape = Location), size = 8) +
scale_shape_manual(values = c("mudflats" = 15, "shore" = 18)) +
scale_color_manual(values = sample_colors) +
geom_smooth(aes(y = predicted), method = "glm", method.args = list(family = "poisson"),
se = TRUE, linetype = 'dotdash', color = "black", linewidth = 0.3) +
labs(color = "Location", shape = "Location", x = "Total read count",
y = "Number of observed ZOTUs") +
theme_bw() +
theme(legend.position = "bottom")
p
p1 <- ggMarginal(p, type = 'densigram')
p1
Call:
glm(formula = total_observations ~ total_read_count, family = poisson(link = "log"),
data = metadata_table)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 2.323e+00 4.321e-01 5.376 7.64e-08 ***
total_read_count -2.640e-05 3.846e-05 -0.686 0.493
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for poisson family taken to be 1)
Null deviance: 10.829 on 10 degrees of freedom
Residual deviance: 10.330 on 9 degrees of freedom
AIC: 56.477
Number of Fisher Scoring iterations: 4
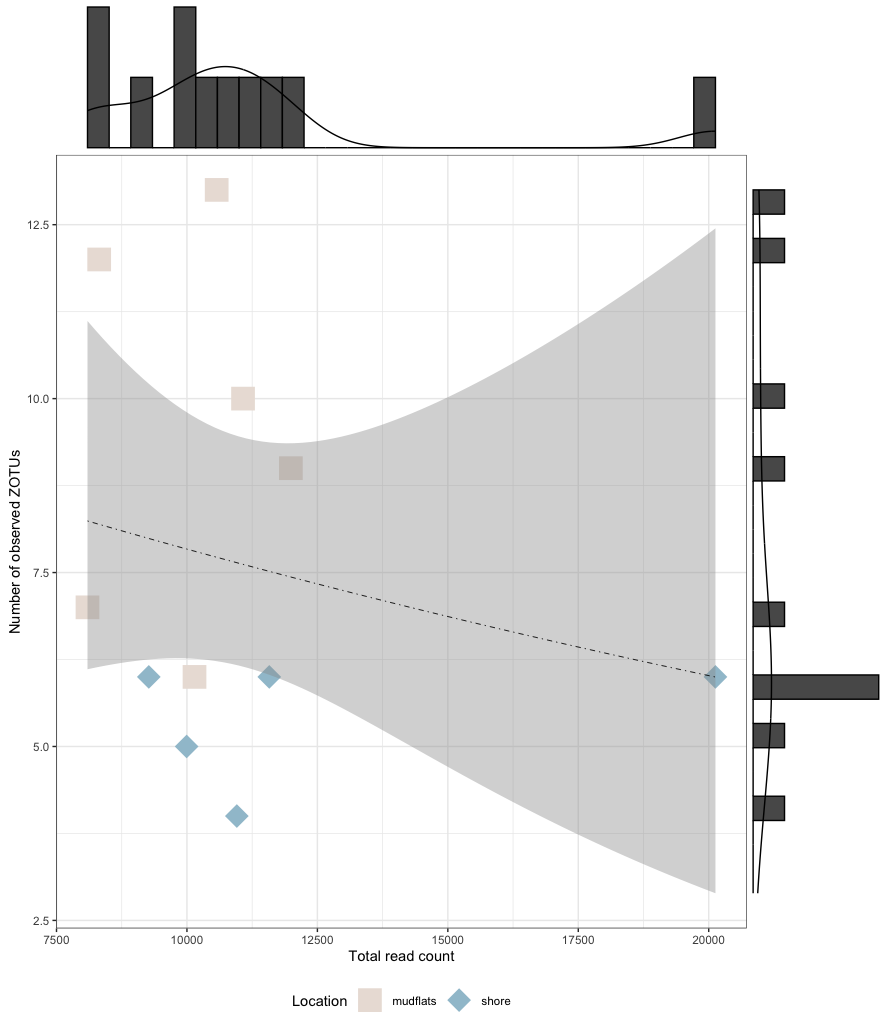
4.3 Rarefaction curves
## read data into R
count_table <- read.table('zotutable_new.txt', header = TRUE, sep = '\t', check.names = FALSE, comment.char = '')
colnames(count_table)[1] <- "V1"
taxonomy_table <- read.table('blast_taxonomy_new.txt', header = FALSE, sep = '\t', check.names = FALSE, comment.char = '')
metadata_table <- read.table('../stats/metadata.txt', header = TRUE, sep = '\t', check.names = FALSE, comment.char = '')
## prepare data for analysis and plotting
count_table <- merge(count_table, taxonomy_table[, c("V1", "V4")], by = "V1", all.x = TRUE)
colnames(count_table)[1] <- "ASV"
colnames(count_table)[ncol(count_table)] <- "Species"
metadata_table$Location <- factor(metadata_table$Location, levels = c("shore", "mudflats"))
## load dataframes into ampvis2 format
ampvis_df <- amp_load(count_table, metadata_table)
## generate rarefaction curves
sample_colors <- c("mudflats" = "#EAE0DA", "shore" = "#A0C3D2")
rarefaction_curves <- amp_rarecurve(ampvis_df, stepsize = 100, color_by = 'Location') +
ylab('Number of observed ZOTUs') +
theme_classic() +
scale_color_manual(values = sample_colors) +
theme(legend.position = "bottom")
rarefaction_curves
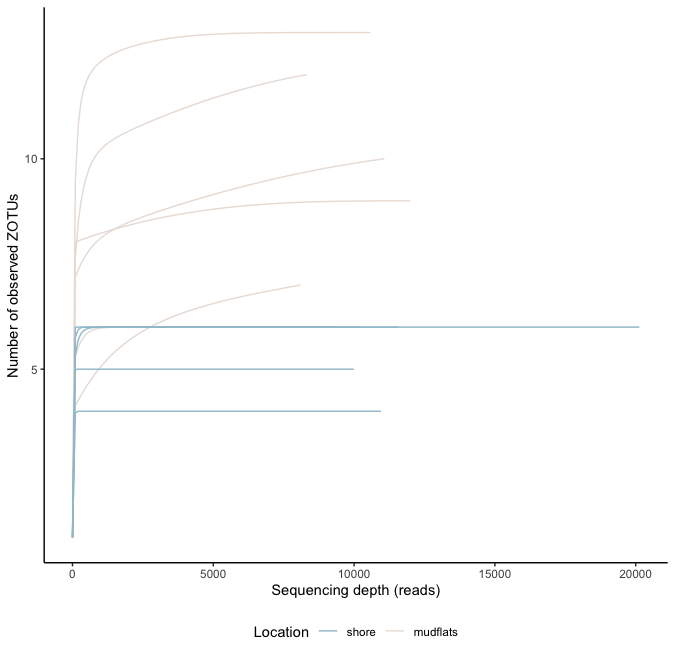
4.4 Curvature indices
## read data in R
count_table <- read.table('zotutable_new.txt', header = TRUE, sep = '\t', check.names = FALSE, comment.char = '')
## create empty dataframe to place curvature indices in
curvature_df <- data.frame("sampleID" = numeric(0), "curvatureIndex" = numeric(0))
## iterate over columns and create new data frames
for (i in 2:ncol(count_table)) {
col1_name <- names(count_table)[1]
col2_name <- names(count_table)[i]
new_df <- data.frame(count_table[, 1], count_table[, i])
names(new_df) <- c(col1_name, col2_name)
## function to generate the rarefaction data from a given sample
dss <- DivSubsamples(new_df, nrf=100, minrarefac=1, NResamples=10)
## calculate curvature index
curvature_value <- Curvature(dss)
# add info to empty dataframe
new_row <- data.frame("sampleID" = col2_name, "curvatureIndex" = curvature_value)
cat("item:", i-1, "/", ncol(count_table), "; sample:", col2_name, "; curvature index: ", curvature_value, "\n")
curvature_df <- rbind(curvature_df, new_row)
}
print(curvature_df)
sampleID curvatureIndex
1 AM1 0.8253815
2 AM2 0.9797766
3 AM3 0.7798546
4 AM4 0.9281280
5 AM5 0.9627029
6 AM6 0.8358680
7 AS2 0.9918151
8 AS3 0.9941130
9 AS4 0.9929947
10 AS5 0.9926441
11 AS6 0.9897328
5. Species accumulation curves
The last data curation method we will cover in this workshop are species accumulation curves, which are used to assess if sufficient sampling was conducted. They work similar to rarefaction curves, but plot the number of detected ZOTUs on the y-axis and number of samples per group on the x-axis. If the curves do not flatten, caution is warranted with the statistical analysis, as more species could have been uncovered with additional sampling in the region.
## read data in R
count_table <- read.table('zotutable_new.txt', header = TRUE, row.names = 1, sep = '\t', check.names = FALSE, comment.char = '')
metadata_table <- read.table('../stats/metadata.txt', header = TRUE, row.names = 1, sep = '\t', check.names = FALSE, comment.char = '')
## transform count_table to presence-absence
count_table[count_table > 0] <- 1
## generate format that can be read into iNEXT.3D (datatype = "incidence_raw" --> list of matrices)
categories <- unique(metadata_table$Location)
matrix_list <- list(data = list())
for (category in categories) {
matrix_list[["data"]][[category]] <- as.matrix(count_table[, rownames(metadata_table[metadata_table$Location == category, ]), drop = FALSE])
}
## run iNEXT3D
inext_out <- iNEXT3D(data = matrix_list$data, diversity = 'TD', q = c(0, 1, 2), datatype = 'incidence_raw', nboot = 50)
## explore results
inext_out$TDInfo
inext_out$TDAsyEst
ggiNEXT3D(inext_out, type = 1, facet.var = 'Assemblage') +
facet_wrap(~Assemblage, nrow = 3) +
theme(title = element_blank(),
panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
panel.spacing = unit(1, "lines"))
estimate3D(matrix_list$data, diversity = 'TD', q = 0, datatype = 'incidence_raw', base = 'coverage', level = 0.9)
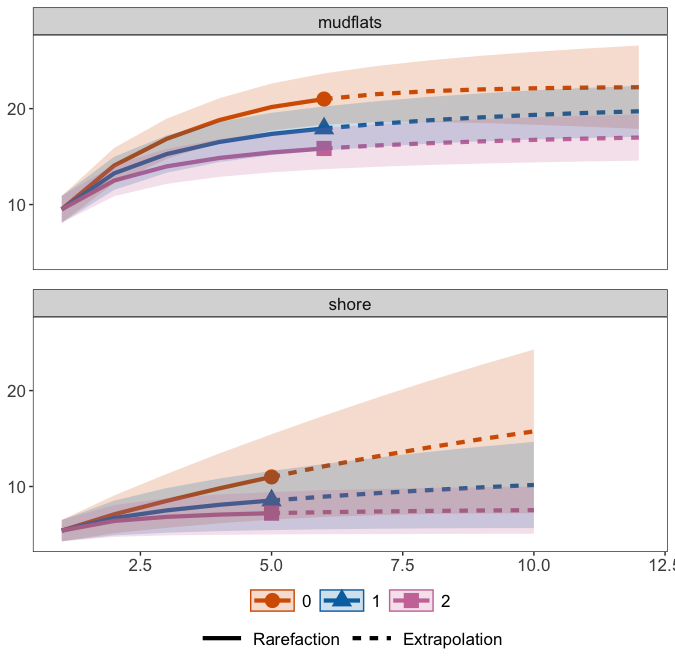
6. Summary
At this point, we have conducted 4 data curation steps, including:
- The removal of spurious artefacts using tombRaider.
- The removal of contaminating signals.
- Identified sequencing coverage was sufficient per sample and determined rarefying the data would not be necessary.
- Identified sufficient sampling for each location
After these steps, we are ready to commence the statistical analysis!
Key Points
The presence of artefacts inflate diversity metrics, but can be removed using novel algorithms
The need for rarefaction can be tested using several methods
Statistics and Graphing with R
Overview
Teaching: 10 min
Exercises: 50 minQuestions
How do I use ggplot and other programs to visualize metabarcoding data?
What statistical approaches do I use to explore my metabarcoding results?
Objectives
Create bar plots of taxonomy and use them to show differences across the metadata
Create distance ordinations and use them to make PCoA plots
Compute species richness differences between sampling locations
Statistical analysis in R
In this last section, we’ll go over a brief introduction on how to analyse the data files you’ve created during the bioinformatic analysis, i.e., the frequency table and the taxonomy assignments. Today, we’ll be using R for the statistical analysis, though other options, such as Python, could be used as well. Both of these two coding languages have their benefits and drawbacks. Because most people know R from their Undergraduate classes, we will stick with this option for now.
Please keep in mind that we will only be giving a brief introduction to the statistical analysis. This is by no means a full workshop into how to analyse metabarcoding data. Furthermore, we’ll be sticking to basic R packages to keep things simple and will not go into dedicated metabarcoding packages, such as Phyloseq.
1. Importing data in R
Before we can analyse our data, we need to import and format the files we created during the bioinformatic analysis, as well as the metadata file. We will also need to load in the necessary R libraries.
## prepare R environment
setwd("~/obss_2024/edna/final")
## add workshop library
.libPaths(c(.libPaths(), "/nesi/project/nesi02659/obss_2024/R_lib"))
## load libraries
library('vegan')
library('lattice')
library('ggplot2')
library('labdsv')
library('tidyr')
library('tibble')
library('reshape2')
library('viridis')
## read data into R
count_table <- read.table('zotutable_new.txt', header = TRUE, sep = '\t', check.names = FALSE, comment.char = '')
taxonomy_table <- read.table('blast_taxonomy_new.txt', header = FALSE, sep = '\t', check.names = FALSE, comment.char = '')
metadata_table <- read.table('../stats/metadata.txt', header = TRUE, sep = '\t', check.names = FALSE, comment.char = '')
## merge taxonomy information with count table
colnames(count_table)[1] <- "V1"
count_table <- merge(count_table, taxonomy_table[, c("V1", "V4")], by = "V1", all.x = TRUE)
count_table$V1 <- NULL
colnames(count_table)[ncol(count_table)] <- "Species"
count_table <- aggregate(. ~ Species, data = count_table, FUN = sum)
rownames(count_table) <- count_table$Species
count_table$Species <- NULL
count_table_pa <- count_table
count_table_pa[count_table_pa > 0] <- 1
2. Preliminary data exploration
One of the first things we would want to look at, is to determine what is contained in our data. We can do this by plotting the abundance of each taxon for every sample in a stacked bar plot.
## plot abundance of taxa in stacked bar graph
ab_info <- tibble::rownames_to_column(count_table, "Species")
ab_melt <- melt(ab_info)
ggplot(ab_melt, aes(fill = Species, y = value, x = variable)) +
geom_bar(position = 'stack', stat = 'identity')
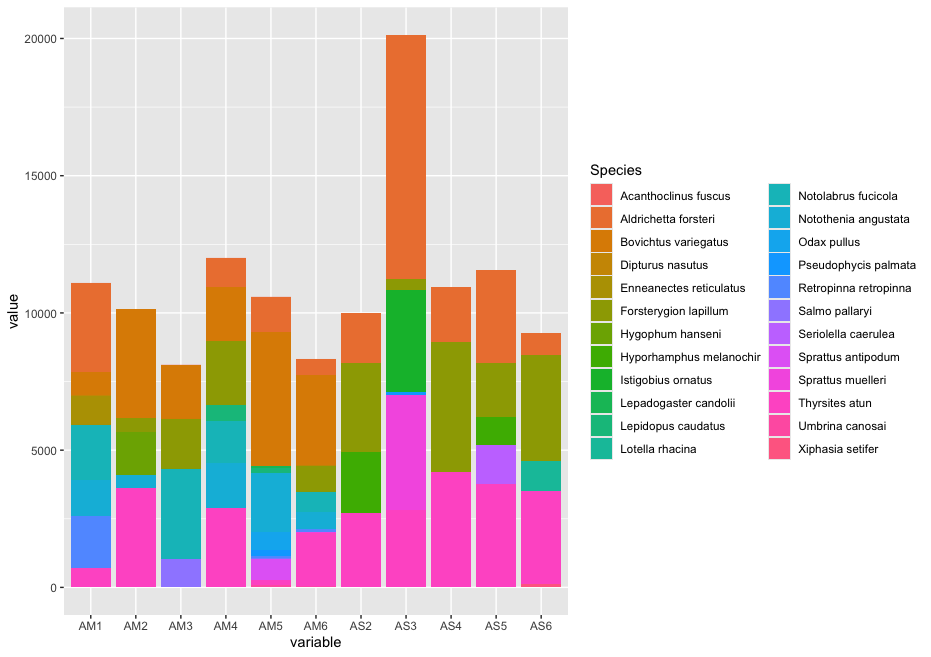
Since the number of sequences differ between samples, it might sometimes be easier to use relative abundances to visualise the data.
## plot relative abundance of taxa in stacked bar graph
ggplot(ab_melt, aes(fill = Species, y = value, x = variable)) +
geom_bar(position="fill", stat="identity")
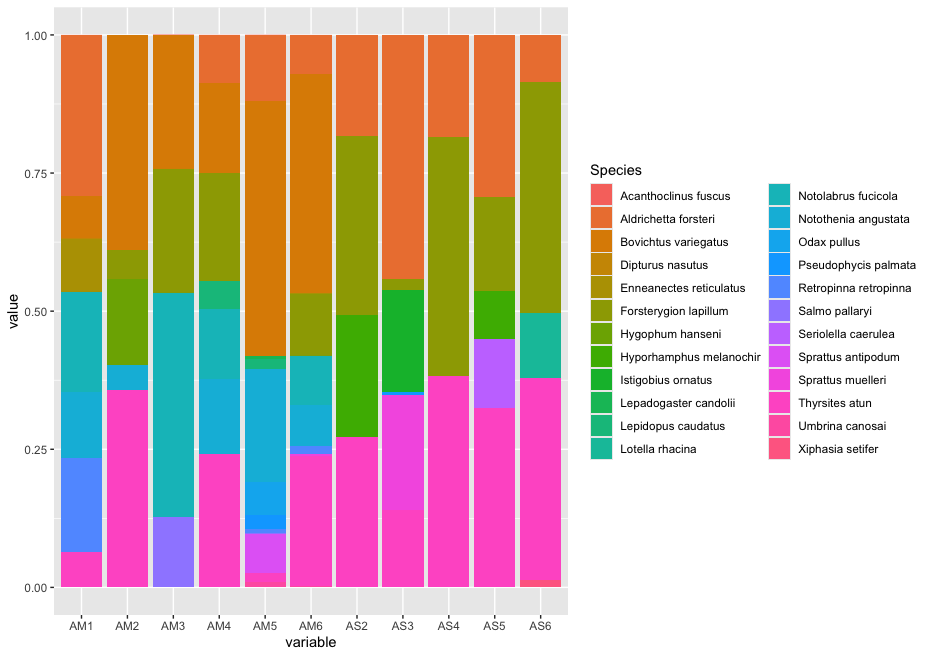
While we can start to see that there are some differences between sampling locations with regards to the observed fish, let’s combine all samples within a sampling location to simplify the graph even further. We can do this by summing or averaging the reads.
# combine samples per sample location and plot abundance and relative abundance of taxa in stacked bar graph
location <- count_table
names(location) <- substring(names(location), 1, 2)
locsum <- t(rowsum(t(location), group = colnames(location), na.rm = TRUE))
locsummelt <- melt(locsum)
ggplot(locsummelt, aes(fill = Var1, y = value, x = Var2)) +
geom_bar(position = 'stack', stat = 'identity')
ggplot(locsummelt, aes(fill = Var1, y = value, x = Var2)) +
geom_bar(position="fill", stat="identity")
locav <- sapply(split.default(location, names(location)), rowMeans, na.rm = TRUE)
locavmelt <- melt(locav)
ggplot(locavmelt, aes(fill = Var1, y = value, x = Var2)) +
geom_bar(position = 'stack', stat = 'identity')
ggplot(locavmelt, aes(fill = Var1, y = value, x = Var2)) +
geom_bar(position="fill", stat="identity")
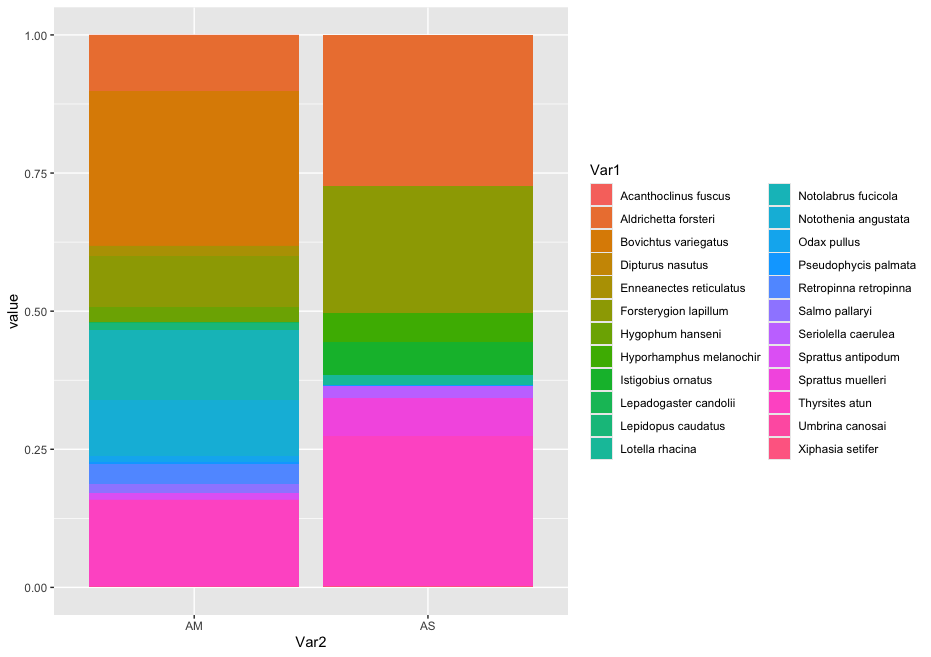
As you can see, the graphs that are produced are easier to interpret, due to the limited number of stacked bars. However, it might still be difficult to identify which species are occurring in only one sampling location, due to the number of species detected and the number of colors used. A trick to use here, is to plot the data per species for the two locations.
## plot per species for easy visualization
ggplot(locsummelt, aes(fill = Var2, y = value, x = Var2)) +
geom_bar(position = 'dodge', stat = 'identity') +
scale_fill_viridis(discrete = T, option = "E") +
ggtitle("Graphing data per species") +
facet_wrap(~Var1) +
theme(legend.position = "non") +
xlab("")
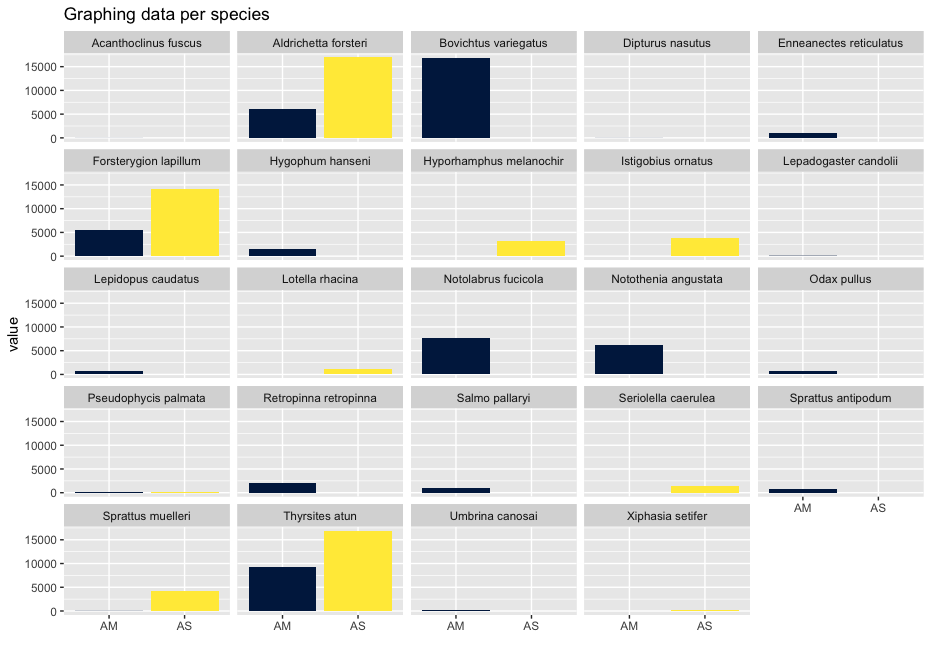
Plotting the relative abundance (transforming it indirectly to presence-absence) could help with distinguishing presence or absence in a sampling location, which could be made difficult due to the potentially big difference in read number.
## plotting per species on relative abundance allows for a quick assessment
## of species occurrence between locations
ggplot(locsummelt, aes(fill = Var2, y = value, x = Var2)) +
geom_bar(position = 'fill', stat = 'identity') +
scale_fill_viridis(discrete = T, option = "E") +
ggtitle("Graphing data per species") +
facet_wrap(~Var1) +
theme(legend.position = "non") +
xlab("")
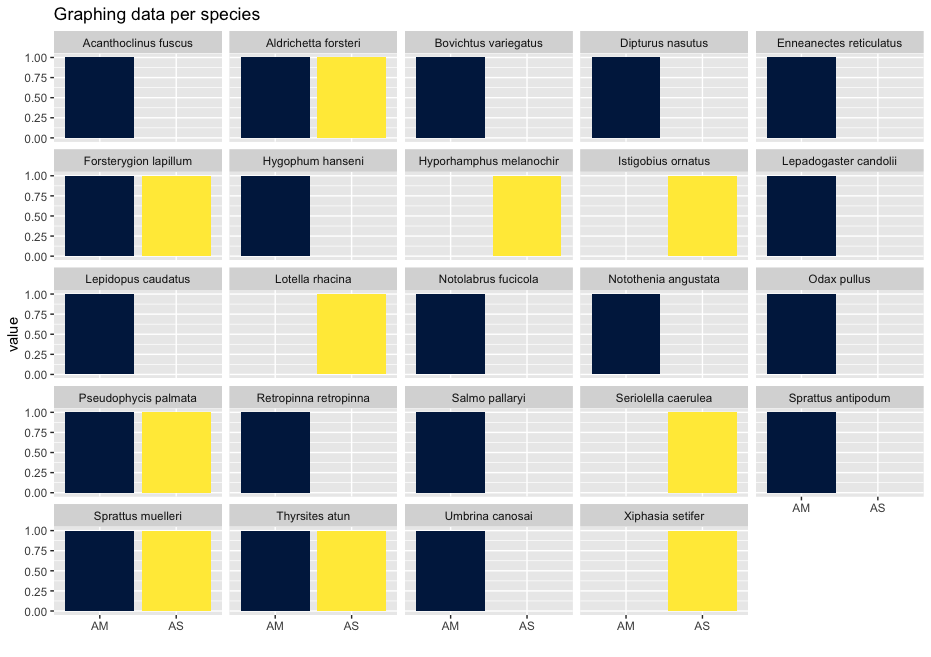
Alpha diversity analysis
Since the correlation between species abudance/biomass and eDNA signal strength has not yet been proven, alpha diversity analysis for metabarcoding data is currently restricted to species richness comparison. In the near future, however, we hope the abundance correlation is proven, enabling more extensive alpha diversity analyses.
## species richness
## calculate number of taxa detected per sample and group per sampling location
specrich <- setNames(nm = c('colname', 'SpeciesRichness'), stack(colSums(count_table_pa))[2:1])
specrich <- transform(specrich, group = substr(colname, 1, 2))
## test assumptions of statistical test, first normal distribution, next homoscedasticity
histogram(~ SpeciesRichness | group, data = specrich, layout = c(1,2))
bartlett.test(SpeciesRichness ~ group, data = specrich)
## due to low sample number per location and non-normal distribution, run Welch's t-test and visualize using boxplot
stat.test <- t.test(SpeciesRichness ~ group, data = specrich, var.equal = FALSE, conf.level = 0.95)
stat.test
boxplot(SpeciesRichness ~ group, data = specrich, names = c('AM', 'AS'), ylab = 'Species Richness')
Welch Two Sample t-test
data: SpeciesRichness by group
t = 3.384, df = 6.5762, p-value = 0.01285
alternative hypothesis: true difference in means between group AM and group AS is not equal to 0
95 percent confidence interval:
1.333471 7.799862
sample estimates:
mean in group AM mean in group AS
9.166667 4.600000
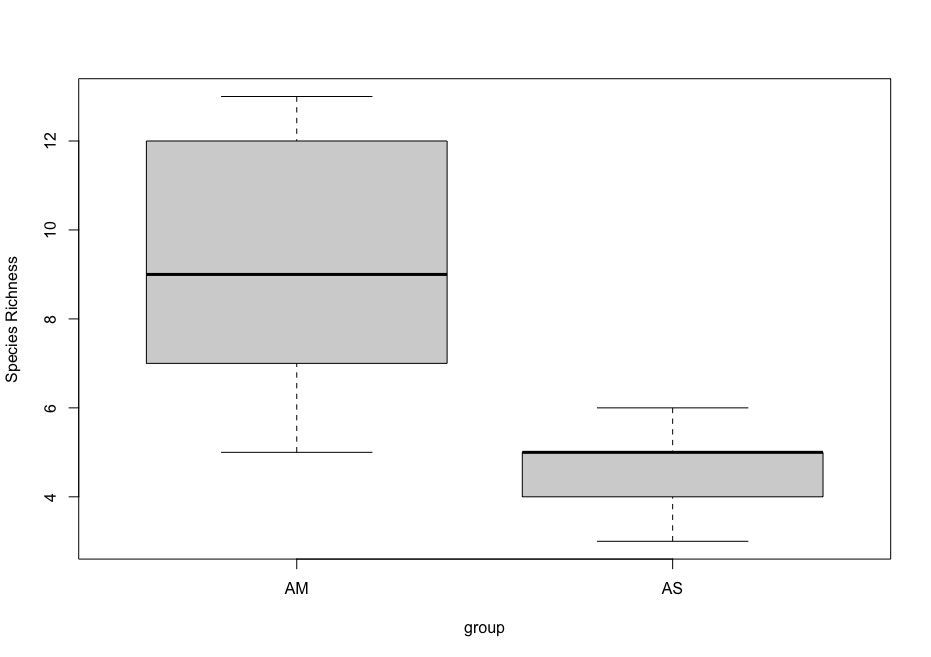
Beta diversity analysis
One final type of analysis we will cover in today’s workshop is in how to determine differences in species composition between sampling locations, i.e., beta diversity analysis. Similar to the alpha diversity analysis, we’ll be working with a presence-absence transformed dataset.
Before we run the statistical analysis, we can visualise differences in species composition through ordination.
## beta diversity
## transpose dataframe
count_table_pa_t <- t(count_table_pa)
dis <- vegdist(count_table_pa_t, method = 'jaccard')
groups <- factor(c(rep(1,6), rep(2,5)), labels = c('mudflats', 'shore'))
dis
groups
mod <- betadisper(dis, groups)
mod
## graph ordination plot and eigenvalues
plot(mod, hull = FALSE, ellipse = TRUE)
ordination_mds <- wcmdscale(dis, eig = TRUE)
ordination_eigen <- ordination_mds$eig
ordination_eigenvalue <- ordination_eigen/sum(ordination_eigen)
ordination_eigen_frame <- data.frame(Inertia = ordination_eigenvalue*100, Axes = c(1:10))
eigenplot <- ggplot(data = ordination_eigen_frame, aes(x = factor(Axes), y = Inertia)) +
geom_bar(stat = "identity") +
scale_y_continuous(expand = c(0,0), limits = c(0, 50)) +
theme_classic() +
xlab("Axes") +
ylab("Inertia %") +
theme(axis.ticks.x = element_blank(), axis.text.x = element_blank())
eigenplot
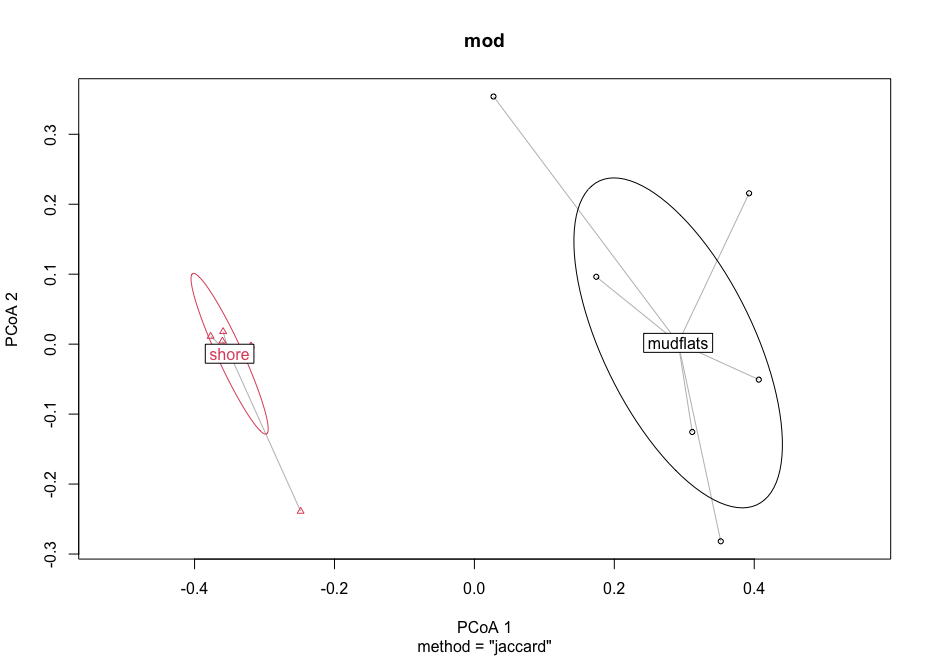
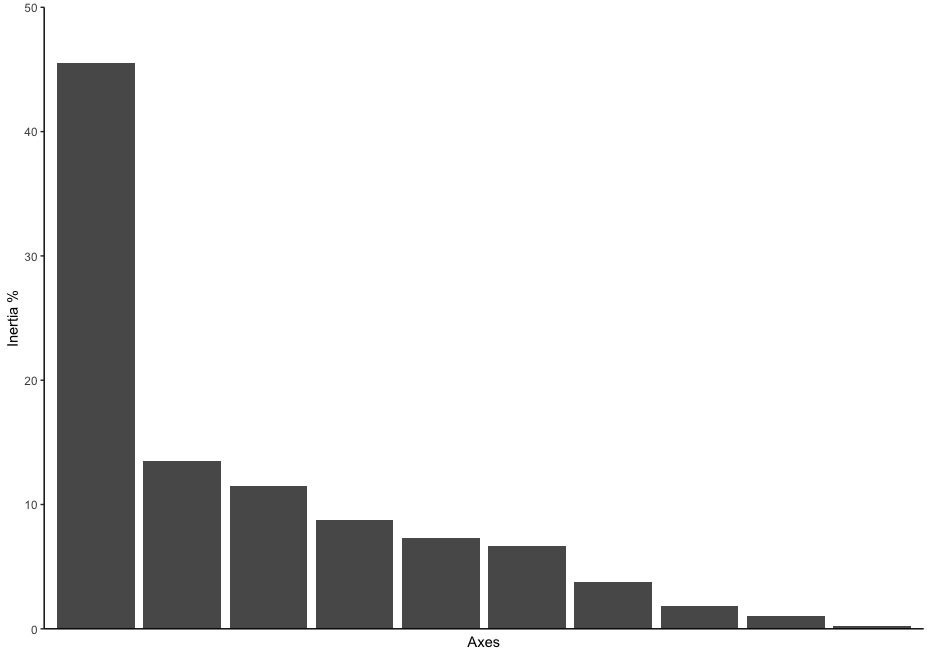
Next, we can run the statistical analysis PERMANOVA to determine if the sampling locations are significantly different. A PERMANOVA analysis will give a significant result when (i) the centroids of the two sampling locations are different and (ii) if the dispersion of samples to the centroid within a sampling location are different to the other sampling location.
## permanova
source <- adonis(count_table_pa_t ~ Location, data = metadata_table, by = 'terms')
print(source$aov.tab)
Permutation: free
Number of permutations: 999
Terms added sequentially (first to last)
Df SumsOfSqs MeanSqs F.Model R2 Pr(>F)
Location 1 0.88031 0.88031 10.146 0.52993 0.001 ***
Residuals 9 0.78087 0.08676 0.47007
Total 10 1.66118 1.00000
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Since PERMANOVA can give significant p-values in both instances, we can run an additional test that only provides a significant p-value if the dispersion of samples to the centroid are different between locations.
## test for dispersion effect
boxplot(mod)
set.seed(25)
permutest(mod)
Permutation test for homogeneity of multivariate dispersions
Permutation: free
Number of permutations: 999
Response: Distances
Df Sum Sq Mean Sq F N.Perm Pr(>F)
Groups 1 0.028786 0.028786 1.4799 999 0.243
Residuals 9 0.175055 0.019451
A final test we will cover today, is to determine which taxa are driving the difference in species composition observed during the PERMANOVA and ordination analyses. To accomplish this, we will calculate indicator values for each taxon and identify differences based on IndValij = Specificityij * Fidelityij * 100.
## determine signals driving difference between sampling locations
specrich$Group <- c(1,1,1,1,1,1,2,2,2,2,2)
ISA <- indval(count_table_pa_t, clustering = as.numeric(specrich$Group))
summary(ISA)
cluster indicator_value probability
Bovichtus.variegatus 1 1.0000 0.003
Acanthoclinus.fuscus 1 0.8333 0.020
Notothenia.angustata 1 0.8333 0.012
Sum of probabilities = 11.46
Sum of Indicator Values = 9.83
Sum of Significant Indicator Values = 2.67
Number of Significant Indicators = 3
Significant Indicator Distribution
1
3
Extra links for further reading:
- https://gjeunen.github.io/hku2023eDNAworkshop/setup.html
- https://nsojournals.onlinelibrary.wiley.com/doi/10.1111/oik.07202
- https://onlinelibrary.wiley.com/doi/full/10.1111/1755-0998.13014
- https://www.biostathandbook.com/onesamplettest.html
- https://rcompanion.org/rcompanion/d_02.html
- https://www.davidzeleny.net/anadat-r/doku.php/en:pcoa_nmds_examples
- https://www.beast2.org/
- https://yulab-smu.top/treedata-book/chapter7.html
That is all for the OBSS 2024 eDNA workshop. If you’d like a more in depth tutorial on eDNA metabarcoding data analysis, please visit my website gjeunen.github.io/hku2023eDNAworkshop. note: Otago Univeristy wifi blocks access, use another connection or a vpn.
Thank you all for your participation. Please feel free to contact me for any questions (gjeunen@gmail.com)!
Key Points
A basic tutorial to conduct statistical analyses of your data