Introduction to the bioinformatic analysis of eDNA metabarcoding data
Overview
Teaching: 15 min
Exercises: 0 minQuestions
What are the main components of metabarcoding pipelines?
Objectives
Review typical metabarcoding workflows
eDNA Workshop
Welcome to the environmental DNA (eDNA) workshop session of the Otago Bioinformatic Spring School 2024!
The primary goal of this course is to introduce you to the analysis of metabarcoding sequence data, with a specific focus on environmental DNA or eDNA, and make you comfortable to explore your own next-generation sequencing data. During this course, we will introduce the concept of metabarcoding and go through the different steps of a bioinformatic pipeline, from raw sequencing data to a count table with taxonomic assignments. At the end of this course, we hope you will be able to assemble paired-end sequences, demultiplex fastq sequencing files, filter sequences based on quality, cluster and denoise sequence data, build your own custom reference database, assign taxonomy to OTUs, further curate you data, and analyse the data in a statistically correct manner.
1. Introduction
Based on the excellent presentations at the start of today’s workshop, we can get an understanding of how broadly applicable eDNA metabarcoding is and how many research fields are using this technology to answer ecological questions.
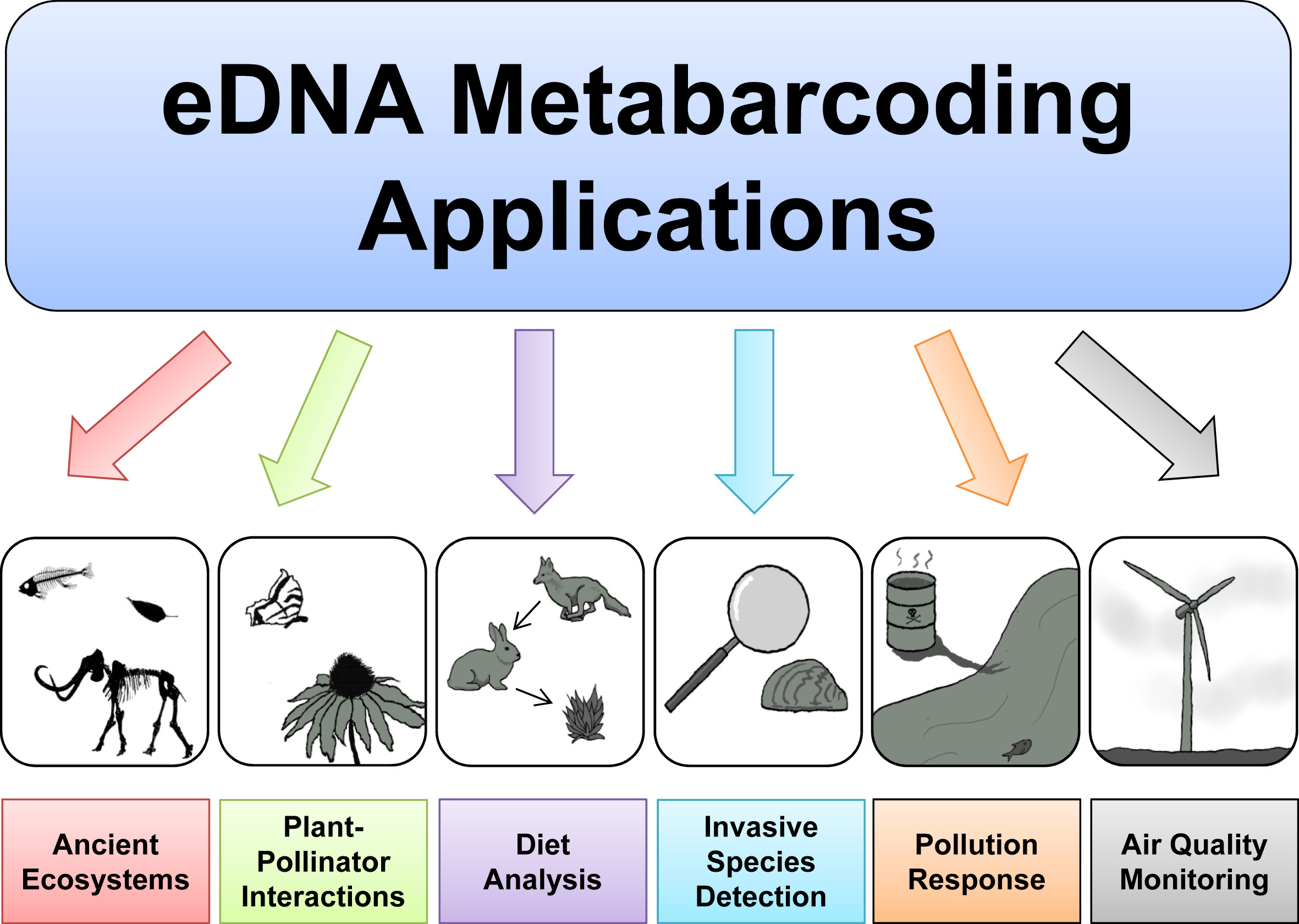
Before we get started with coding, we will introduce the dataset that we will be analysing today, some of the software programs available (including the ones we will use today), and the general structure of a bioinformatic pipeline to process metabarcoding data. Finally, we will go over the folder structure we suggest you use during the bioinformatic processing of metabarcoding data to help keep an overview of the files and ensure reproducibility of the bioinformatic pipeline.
2. Experimental setup
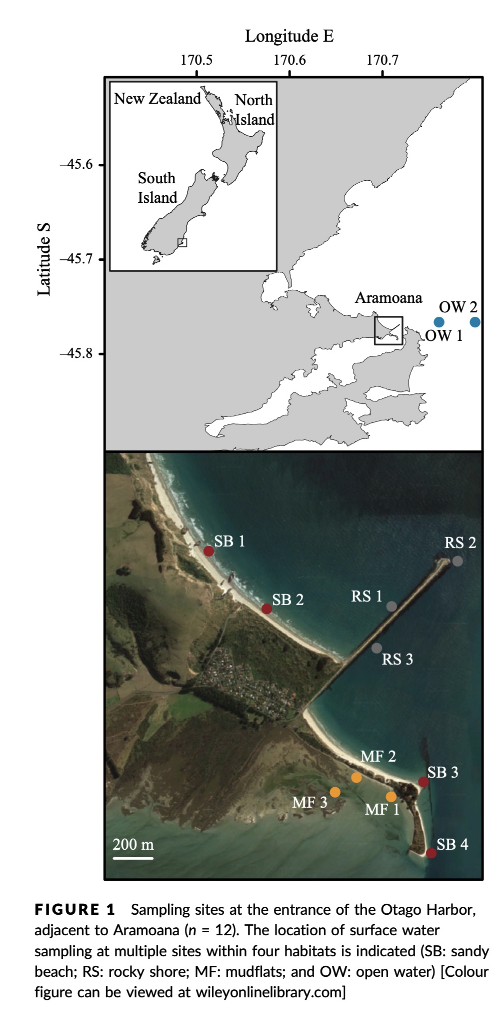
The data we will be analysing today is taken from an experiment we published in 2018 in Molecular Ecology Resources. In this experiment we wanted to determine the spatial resolution of aquatic eDNA in the marine environment. To investigate the spatial resolution, we visited a marine environment with high dynamic water movement where we knew from traditional monitoring that there were multiple habitats present on a small spatial scale containing different community assemblages. Our hypothesis was as follows, if the spatial resolution of aquatic eDNA is high, we would encounter distinct eDNA signals at each habitat resembling the residing community. However, if aquatic eDNA is transported through water movement, from currents and tidal waves, similar eDNA signals would be obtained across this small sampling area, which would be indicative of a low spatial resolution for aquatic eDNA. Our experiment consisted of five replicate water samples (2l) at each sampling point. Water was filtered using a vacuum pump and extracted using a standard Qiagen DNeasy Blood & Tissue Kit. All samples were amplified using three metabarcoding primer sets to target fish and crustacean diversity, specifically. We also used a generic COI primer set to amplify eukaryotic eDNA signals.
For today’s data analysis, we subsetted the data to only include two sampling sites and we’ll only be looking at the fish sequencing data. The reason for doing so, is to ensure all bioinformatic steps will take maximum a couple of minutes, rather than hours or days to complete the full analysis.

3. A note on software programs
The popularity of metabarcoding research, both in the bacterial and eukaryote kingdom, has resulted in the development of a myriad of software packages and pipelines. Furthermore, software packages are continuously updated with new features, as well as novel software programs being designed and published. We will be using several software programs and R packages during this workshop. However, we would like to stress that the pipeline used in the workshop is by no means better than other pipelines and we urge you to explore alternative software programs and trial them with your own data after this workshop.
Metabarcoding software programs can generally be split up into two categories, including:
- stand-alone programs that are self-contained: These programs usually incorporate novel functions or alterations on already-existing functions. Programs can be developed for a specific task within the bioinformatic pipeline or can execute multiple or all steps in the bioinformatic pipeline. Examples of such programs are: Mothur, USEARCH, VSEARCH, cutadapt, dada2, OBITools3.
- wrappers around existing software: These programs attempt to provide an ecosystem for the user to complete all steps within the bioinformatic pipeline without needing to reformat documents or change the coding language. Examples of such programs are: QIIME2 and JAMP.
Two notes of caution about software programs:
- While some software programs are only able to complete a specific aspect of the bioinformatic pipeline, others are able to handle all steps. It might therefore seem easier to use a program that can be used to run through the pipeline in one go. However, this could lead to a ‘black box’ situation and can limit you when other software programs might have additional options or better alternatives. It is, therefore, recommended to understand each step in the bioinformatic pipeline and how different software programs implement these steps. Something we hope you will take away from today’s eDNA workshop.
- One issue you will run into when implementing several software programs in your bioinformatic pipeline is that each program might change the structure of the files in a specific manner. These modifications might lead to incompatability issues when switching fto the next program. It is, therefore, important to understand how a specific program changes the structure of the sequencing file and learn how these modification can be reverted back (through simple python or bash scripts). We will talk a little bit more about file structure and conversion throughout the workshop.
Below, we can find a list of some of the software programs that are available for analysing metabarcoding data (those marked with a ‘*’ will be covered in this workshop):
-
FastQC* (webpage) is a program that assesses the quality of .fastq files.
-
MultiQC (webpage) is a program that combines multple FastQC reports into a single report.
-
Cutadapt* (webpage) is a program that finds and removes adapter sequences, primers, and other types of unwated sequences from high-throughput sequencing reads. We will use this program to demultiplex our data and remove the primer-binding regions.
-
USEARCH/VSEARCH* (USEARCH), and the open-source version (which we will use) (VSEARCH*), has many utilities for running metabarcoding analyses. Many of the most common pipelines use at least some of the tools from these packages.
-
CRABS* (webpage) is a software package developed by our team that lets you build and curate custom reference databases.
-
tombRaider* (webpage) is an algorithm developed by our team that identifies and removes artefacts from metabarcoding datasets.
-
Qiime2 is actually a collection of different programs and packages, all compiled into a single ecosystem so that all things can be worked together. On the Qiime2 webpage there is a wide range of tutorials and help.
-
Dada2 is a good program, run in the R language. There is good documentation and tutorials for this on its webpage. You can actually run Dada2 within Qiime, but there are a few more options when running it alone in R.
-
Mothur has long been a mainstay of metabarcoding analyses, and is worth a look
-
Another major program for metabarcoding is OBITools, which was one of the first to specialise in non-bacterial metabarcoding datasets. The latest version is built using the coding language GO and is currently in Beta. Check the main webpage to keep track of its development.
Additionally, there are several good R packages that can be used to analyse and graph results. Chief among these are Phyloseq, iNEXT.3D, ape, microbiome, indicspecies, vegan, and DECIPHER.
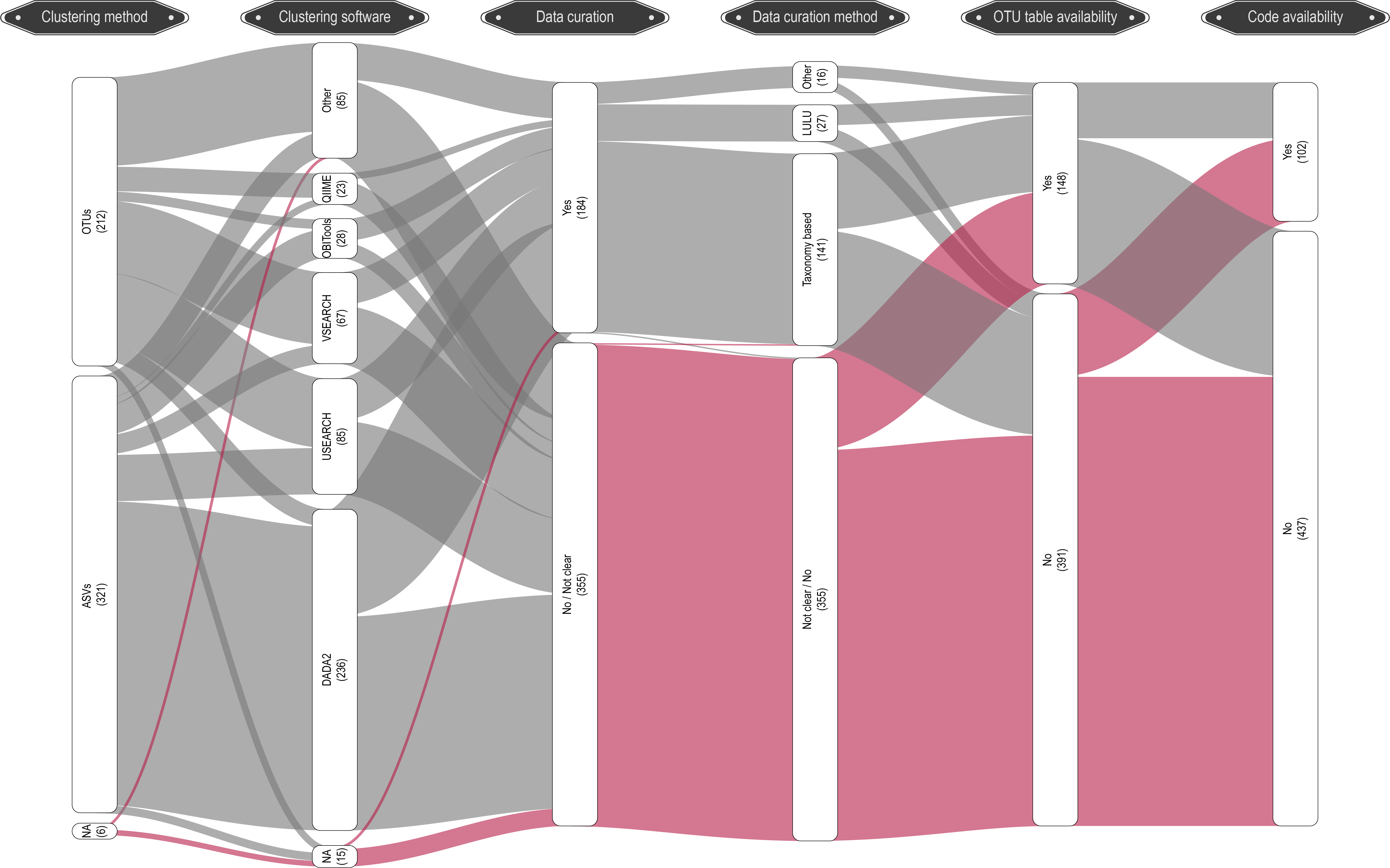
In a recent meta-analysis our lab conducted, we can see that DADA2 and USEARCH/VSEARCH are the most prominently used software programs in the metabarcoding research community.
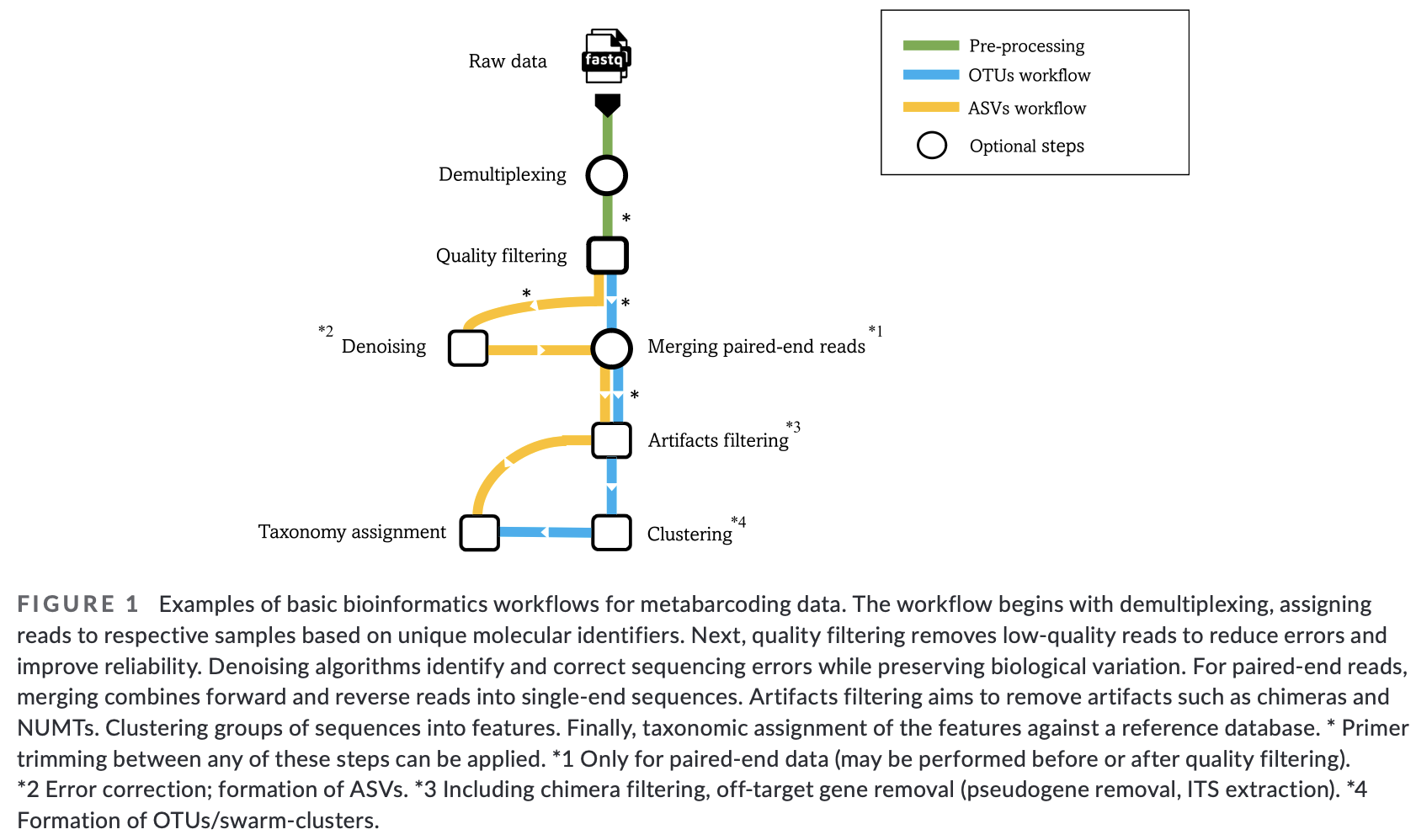
4. General overview of the bioinformatic pipeline
Although software options are numerous, each pipeline follows more or less the same basic steps and accomplishes these steps using similar tools. Before we get started with coding, let’s quickly go over the main steps of the pipeline to give you a clear overview of what we will cover today. For each of these steps, we will provide more information when we cover those sections during the workshop.
5. A note on scripts and reproducible code
One final topic we need to cover before we get started with coding is the use of scripts to execute code. During this workshop, we will create very small and simple scripts that contain the code used to process the sequencing data. While it is possible to run the commands without using scripts, it is good practice to use this method for the following three reasons:
- By using scripts to execute the code, there is a written record of how the data was processed. This will make it easier to remember how the data was processed in the future.
- While the sample data provided in this tutorial is small and computing steps take up a maximum of several minutes, processing large data sets can be time consuming. It is, therefore, recommended to process a small portion of the data first to test the code, modify the filenames once everything is set up, and run it on the full data set.
- Scripts can be reused in the future by changing file names, saving you time by not having to constantly write every line of code in the terminal. Minimising the amount needing to be written in the terminal will, also, minimise the risk of receiving error messages due to typo’s.
6. The folder structure
That is it for the intro, let’s get started with coding!
First, we will have a look at the folder structure that will be used during the workshop. For any project, it is important to organise your various files into subfolders. Through the course you will be navigating between folders to run your analyses. It may seem confusing or tedious at first, especially as there are only a few files we will be dealing with during this workshop. However, keep in mind that for many projects you can easily generate hundreds of files, so it is best to start with good practices from the beginning.
Let’s navigate to the edna folder in the ~/obss_2024/ directory. Running the ls -ltr command outputs the following subfolders:
- raw/
- input/
- output/
- meta/
- scripts/
- final/
- refdb/
- stats/
Most of these are currently empty, but by the end of the course they will be full of the results of your analysis.
So, let’s begin!
Key Points
Most metabarcoding pipelines include the same basic components
Frequency tables are key to metabarcoding analyses