Nanopore-based sequencing: introduction
Overview
Teaching: 35 min
Exercises: 0 minQuestions
What is nanopore-based sequencing?
Objectives
Understand details of ONT sequencing platforms.
What is this module about?
- Long-read sequencing with the Oxford Nanopore Technologies (ONT) platform.
- WHY?
- recent technology advances have led to a surge in popularly for this platform.
- Huge scope for really exciting (and low cost) science
- Concepts and skills learned here are transferable to other sequencing technologies.
- WHAT?
- Genome assembly
- RNA-seq
- Variant identification (single base & structural)
- Base modification (e.g., methylation)
- Metagenomics
ONT platforms
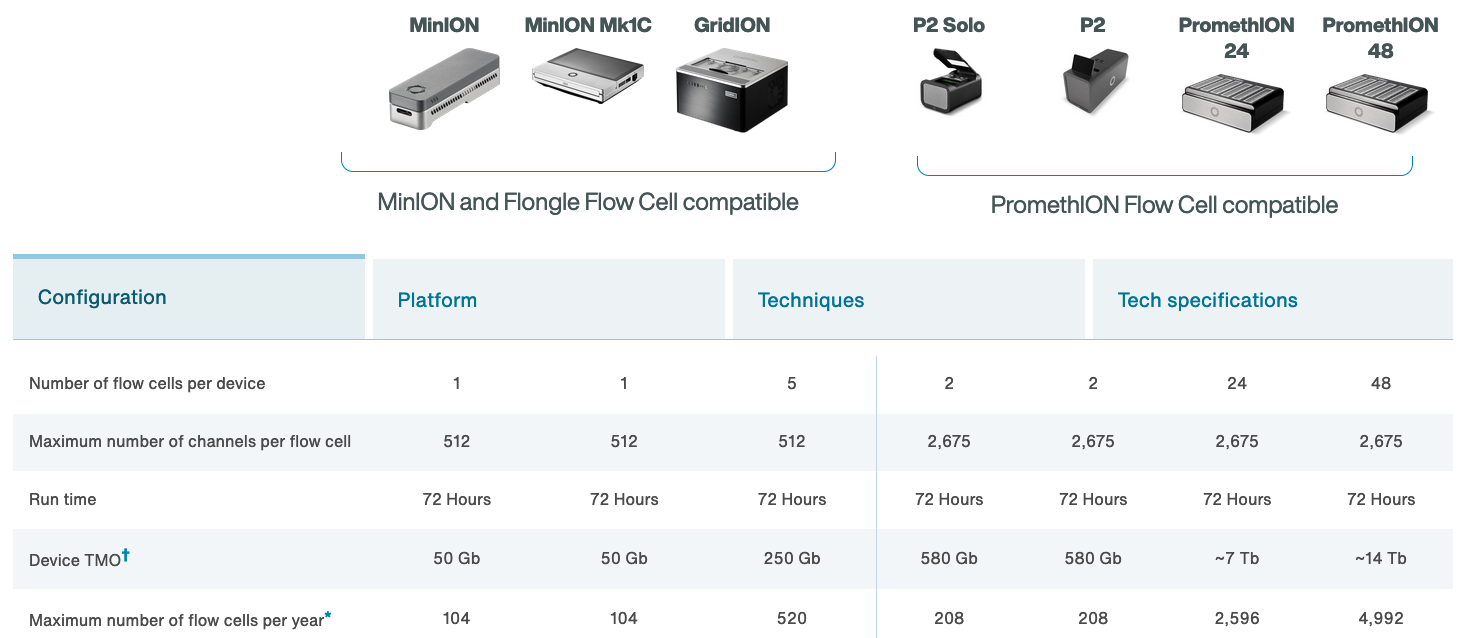
The MinION
http://www.nature.com/news/data-from-pocket-sized-genome-sequencer-unveiled-1.14724
- Rough specs (2014)
- 6-8 hour run time
- sequence per run: ~110Mbp
- average read length: 5,400bp
- reads up to 10kbp
- 2023 specs:
- can run for up to 72 hours
- Maximum (theoretical) yield per run: 50Gbp
- Maximum read length recorded: >4Mbp

https://nanoporetech.com https://nanoporetech.com/products/specifications
The MinION Mk1B
- The MinION Mk1B is the current version of ONT’s original sequencer.
- Connects to a computer via USB.

https://nanoporetech.com/products/minion

The MinION Mk1C
- The MinION Mk1C provides a truly portable sequencing option, with built in compute and touchscreen.
- For the price, however, the hardware is not particularly impressive.

https://nanoporetech.com/products/minion

The MinION Mk1D
- The next iteration of portable sequencing devices is the Mk1D.
- Tablet-based (plug-in).
- Currently details are limited, but developer versions
arewere scheduled to be released during 2023: “Further details on specifications will be provided in 2023.”

https://nanoporetech.com/products/minion-mk1d
The GridION
- The GridION offers a “medium-throughput” option for Nanopore-based sequencing: can run up to five flowcells at once.
- The MinION (Mk1B and Mk1C) and GridION use the same flow cells
- The Otago Genomics Facility has one of these.

https://nanoporetech.com/products/gridion
The Flongle
- The Flongle uses an adapter to allow a smaller (and cheaper) flow cell to be used in the MinION and GridION devices.
- Single-use system provides low-cost option for targeted sequencing (e.g., diagnostic applications).

https://nanoporetech.com/products/flongle
PromethION
- The higher throughout PromethION uses a smaller cartridge-like flow cell. Two options: 24 or 48 flow cells.

https://nanoporetech.com/products/promethion
A “mini-PromethION”: the P2

Smaller device (standalone or connect to host computer) that can run Promethion flow cells.
https://nanoporetech.com/products/p2
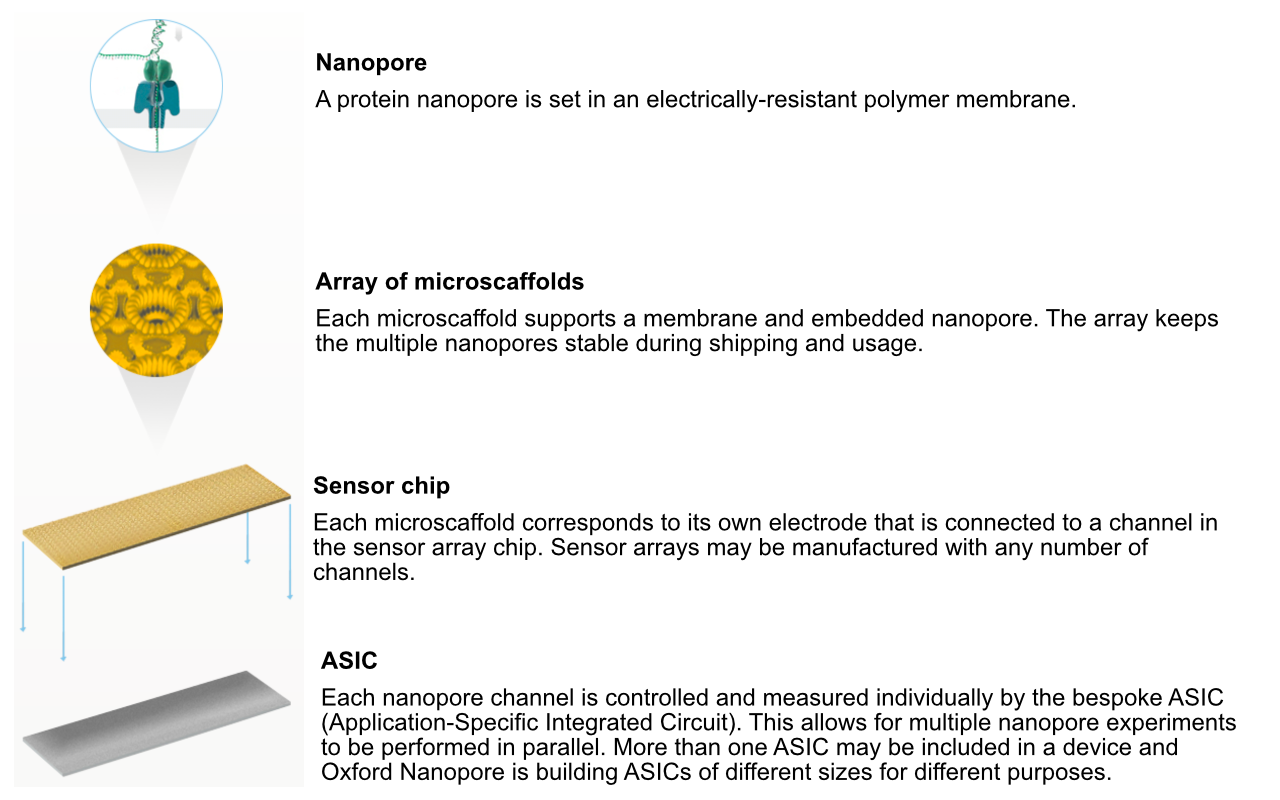
Flowcell characteristics
- Pores are arranged in sets of four to form a “channel”. Periodically during the sequencing run, the ONT software decides which of the four pores to use from each channel (called a “mux scan”).
- MinION flowcells have 512 channels, so 2048 pores: typically ~1200-1800 are “active” (usable pre-run) (ONT guarantees at least 800 active pores).
- PromethION flowcells have 2675 channels, so 10,700 pores (ONT guarantees at least 5000 active pores).
- Sequencing occurs at roughly 450 bases per second (ONT recommends keeping speed above 300 bases per second - additional reagents can be added to “refuel” the flowcell*), although other speeds are now possible (can prioritise data volume vs accuracy).
-
NOTE: pores are not constantly active, and can become blocked during the run
- https://community.nanoporetech.com/protocols/experiment-companion-minknow/v/mke_1013_v1_revbm_11apr2016/refuelling-your-flow-cell
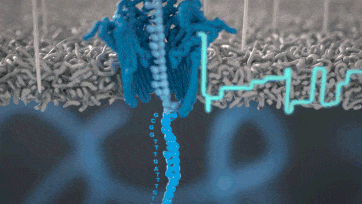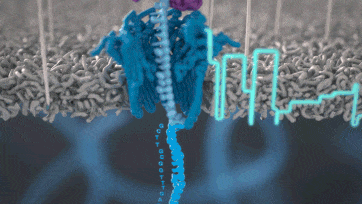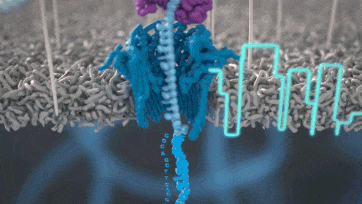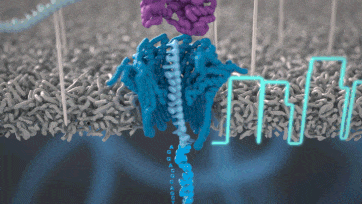
Nanopore technology
https://nanoporetech.com/how-it-works
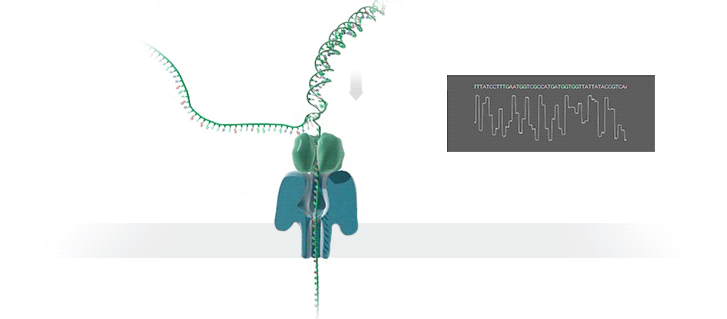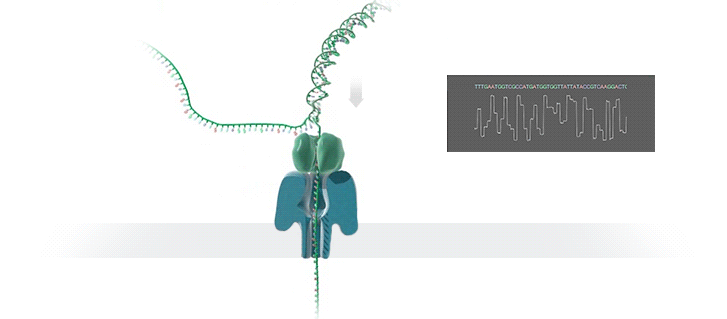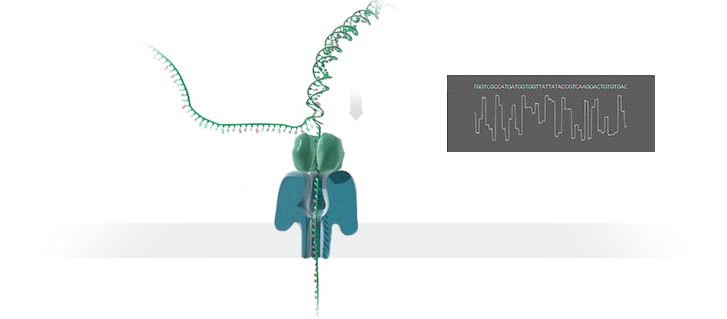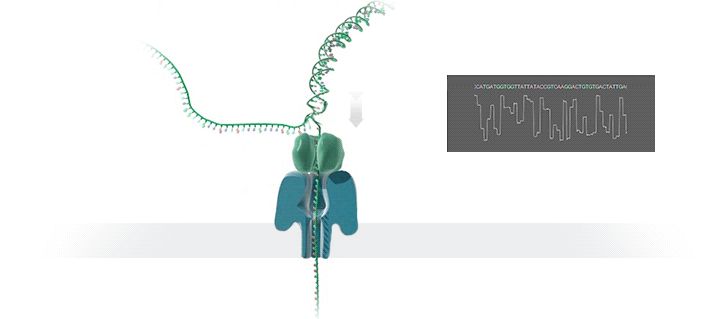
Nanopore technology
- A motor protein (green) passes a strand of DNA through a nanopore (blue). The current is changed as the bases G, A, T and C pass through the pore in different combinations.
https://nanoporetech.com/how-it-works
Nanopore movies
For more detailed information about ONT sequencing:
Technological advances…
- Since its introduction, nanopore sequencing has seen a number of improvements.
- The initial product was realtively slow, expensive (per base sequenced) and error prone (i.e., incorrect bases calls).
- Incremental improvements have led to major advances in both speed and accuracy.
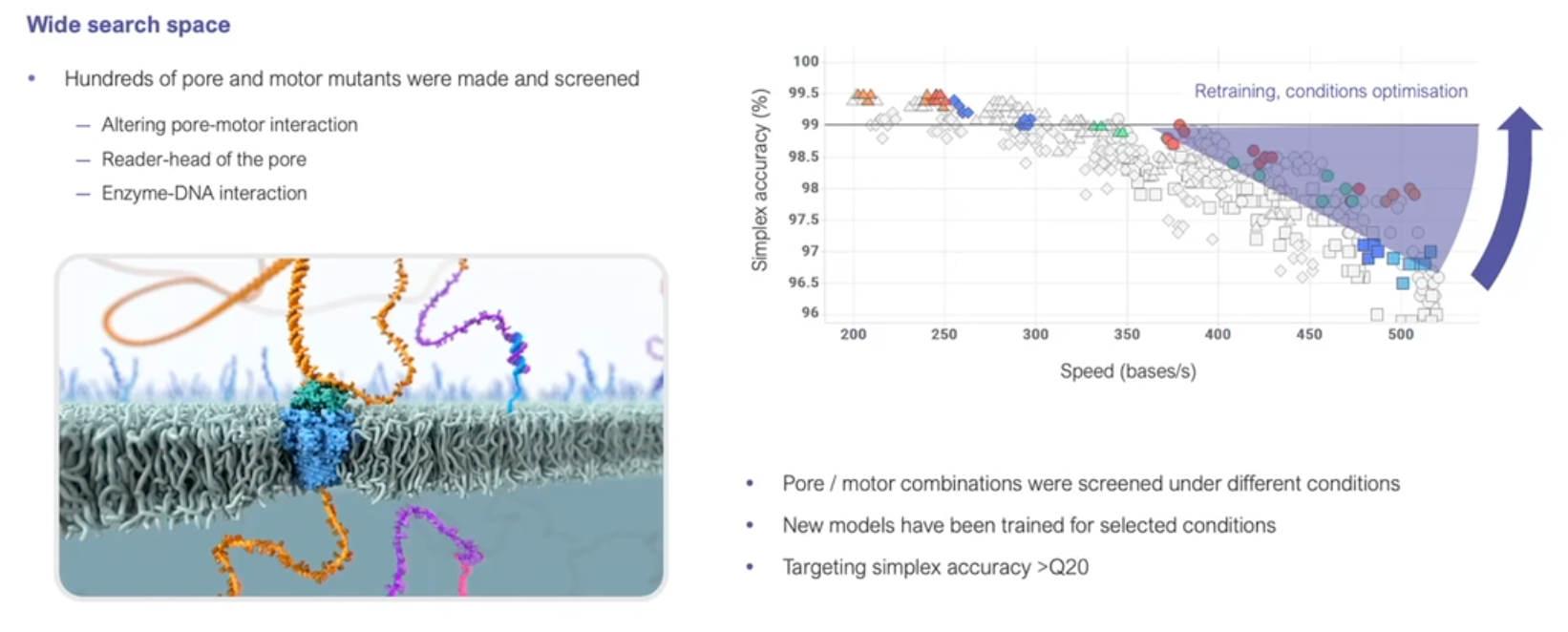
https://nanoporetech.com/resource-centre/london-calling-2022-update-oxford-nanopore-technologies
2D sequencing (prior to 2017)
- Hairpin-based approach provided natural error detection methodology:
- Link DNA strands with a hairpin adapter.
- Sequence template strand followed by complement.
- Basecall and compare sequences to produce consensus.
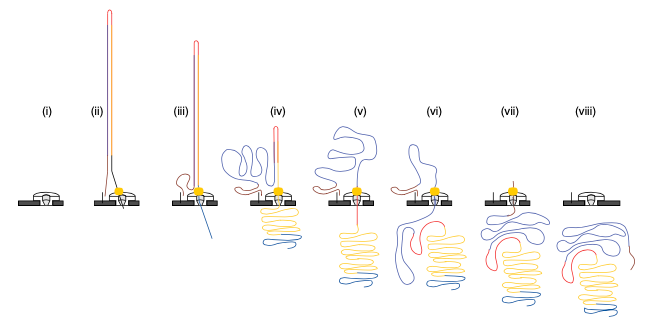
Jain, et al. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol 17, 239 (2016). https://doi.org/10.1186/s13059-016-1103-0
What happened to 2D reads?
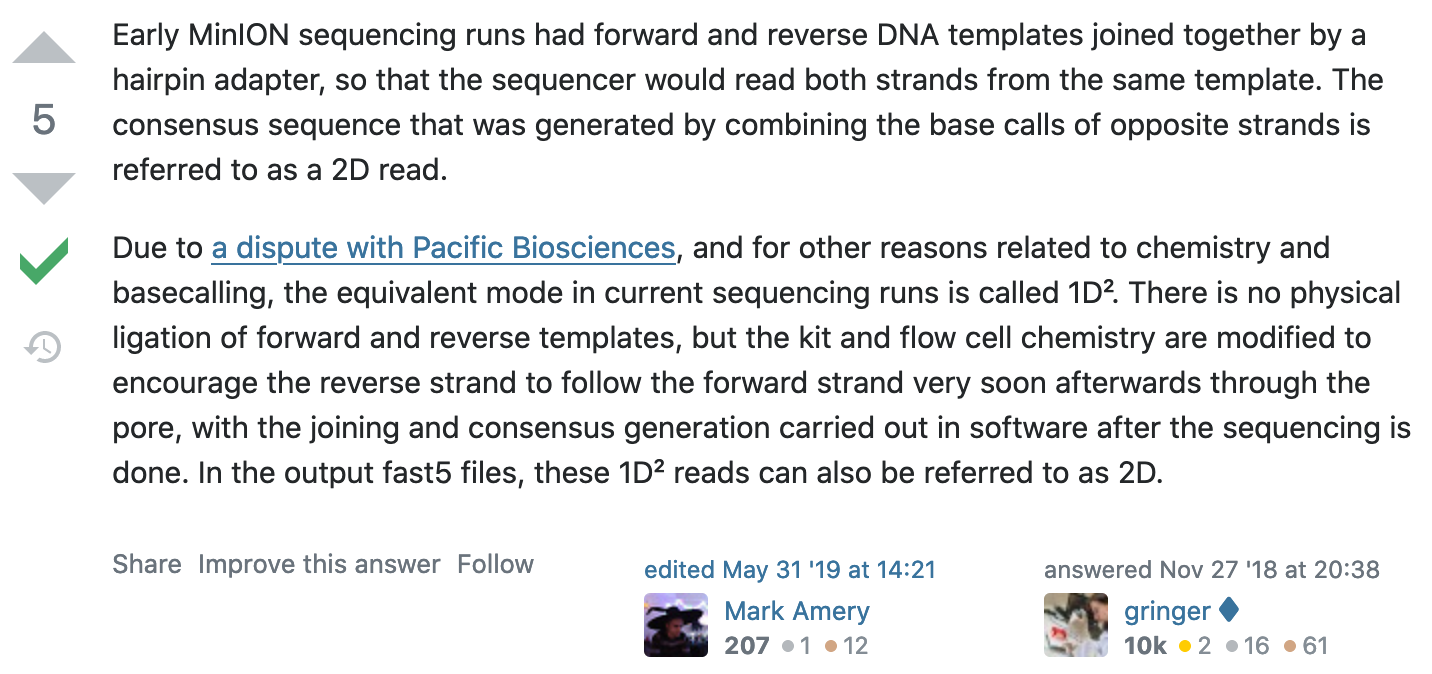
https://bioinformatics.stackexchange.com/questions/5525/what-are-2d-reads-in-the-oxford-minion/5528
The hairpin lawsuit…
- PacBio (competitor in the long-read space) and ONT have filed a number of lawsuits against each other over the past few years.

https://www.genomeweb.com/sequencing/pacbio-oxford-nanopore-settle-patent-dispute-europe
1D2 sequencing
- In 2017 ONT announced the new 1D2 chemistry
- Showed higher accuracy that 1D (and SAID it was better than 2D)
- It didn’t last long…
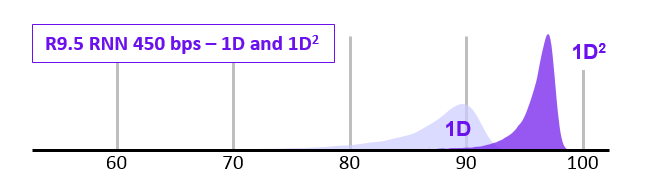
- Video at link below:
https://nanoporetech.com/about-us/news/1d-squared-kit-available-store-boost-accuracy-simple-prep
Pore imporvements: the R10 pore
- ONT introduced the new R10 pore in 2019 (previous was R9.4.1).
- Main differences were longer barrel and dual reader head: gave improved resolution of homopolymer runs.
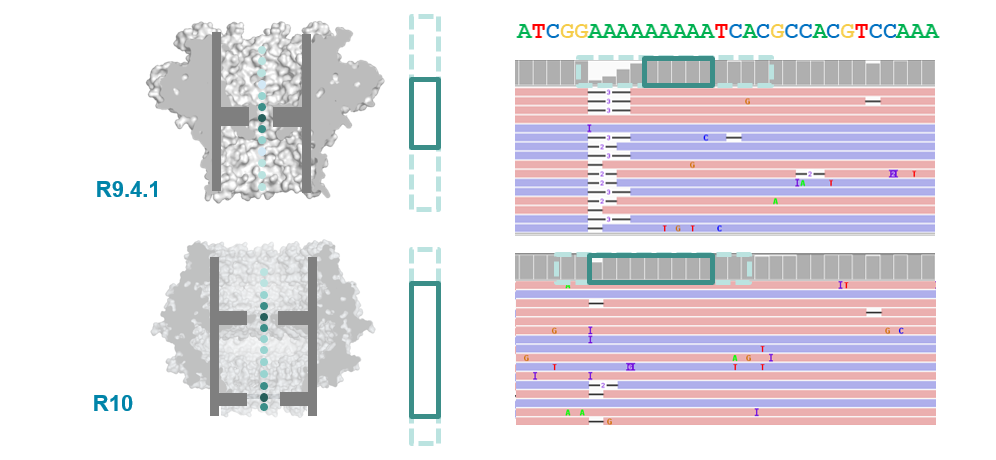
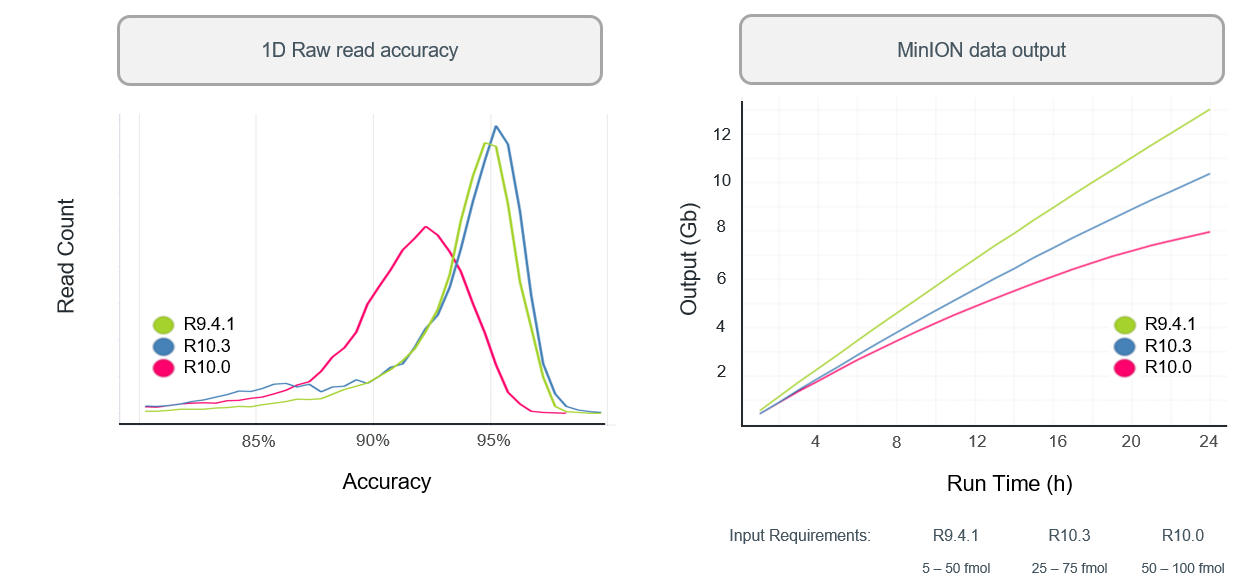
R10.3 vs R9.4.1 performance
- With a bit more tweaking (to get to R10.3) ONT improved 1D (i.e., single-strand) sequencing accuracy, although throughput is still not as high as the R.9.4.1 pore.
Q20+: the return of 2D reads…
- The previous ONT products are “Q10” (we’ll discuss this soon), meaning that the error rate is roughly 1 incorrect base call per 10 bases (that’s high!)
- The new “Q20+” products are now available - moves to less than 1 error per 100 bases (a little more respectable, but still well below short-read technologies like Illumina).
- Upgrade includes a return to the “2D” approach.

Nanopore workflow
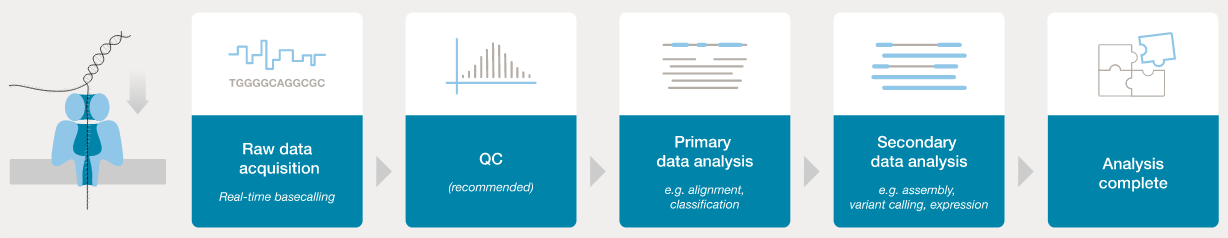
- ONT provides software (MinKNOW) for operating the MinION, and for generating the sequence data (e.g., the
guppyanddoradobasecallers). - Once the raw (POD5 or FAST5 - see next section) data have been converted to basecalls, we can use more familiar tools for quality assessment and analysis (e.g., FastQC).
https://nanoporetech.com/nanopore-sequencing-data-analysis
Basecalling: guppy (deprecated)
guppyis a neural network based basecaller.- analyses the electrical trace data and predicts base
- it is GPU-aware, and can basecall in real time
- can also call base modifications (e.g., 5mC, 6mA)
- high accuracy (HAC) mode (slower) and super-high accuracy (SUP) mode (even slower) can improve basecalls post-sequencing
- MANY other machine learning basecallers have been proposed.
- Output is the standard “FASTQ” format for sequence data.
guppyhas now been “retired” by ONT, and replaced withdorado.
Basecalling: dorado
- ONT has recently released a new base caller:
dorado - Optimised for 10.4.1 flowcells (but there are basecalling models for 9.4.1)
- Designed to support Apple GPUs (M1/M2/M3)
- Like Guppy, can call base modifications
- Also has FAST, HAC and SUP modes for basecalling.
10.4.1 flowcells + v14 chemistry: accuracy
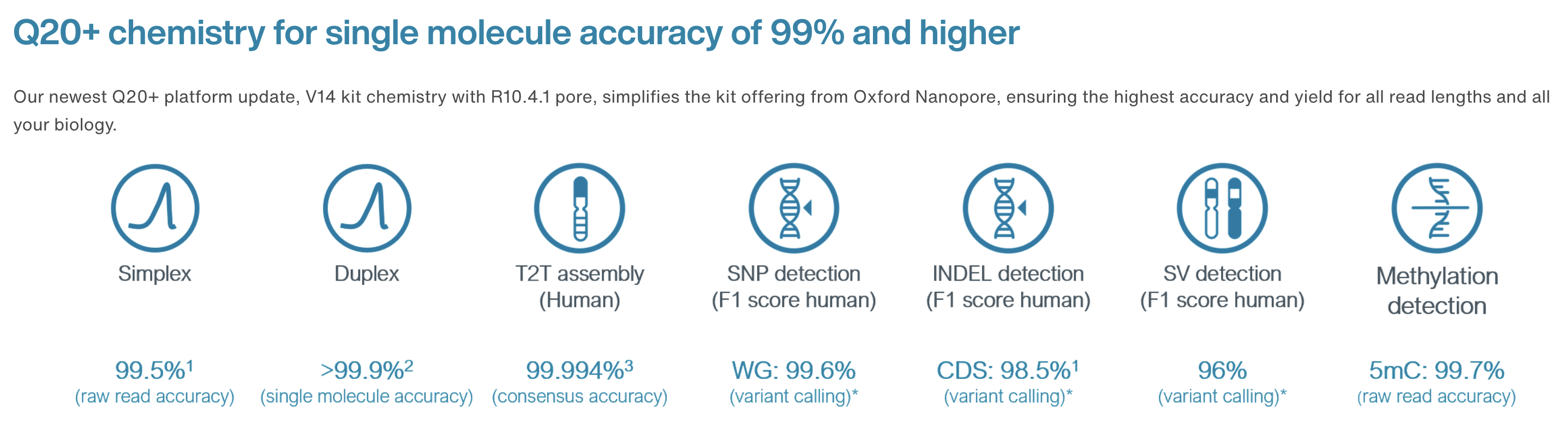
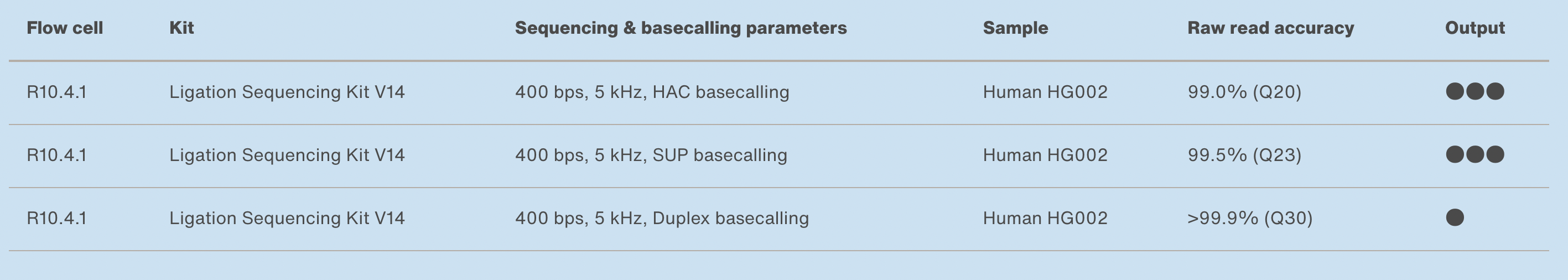
https://nanoporetech.com/accuracy
10.4.1 flowcells + v14 chemistry: simplex
- The new chemistry (v14) and updated flowcells (10.4.1) have moved the quality up to ON AVERAGE 1 error per 100 bases (Q20) for simplex reads (single strand).
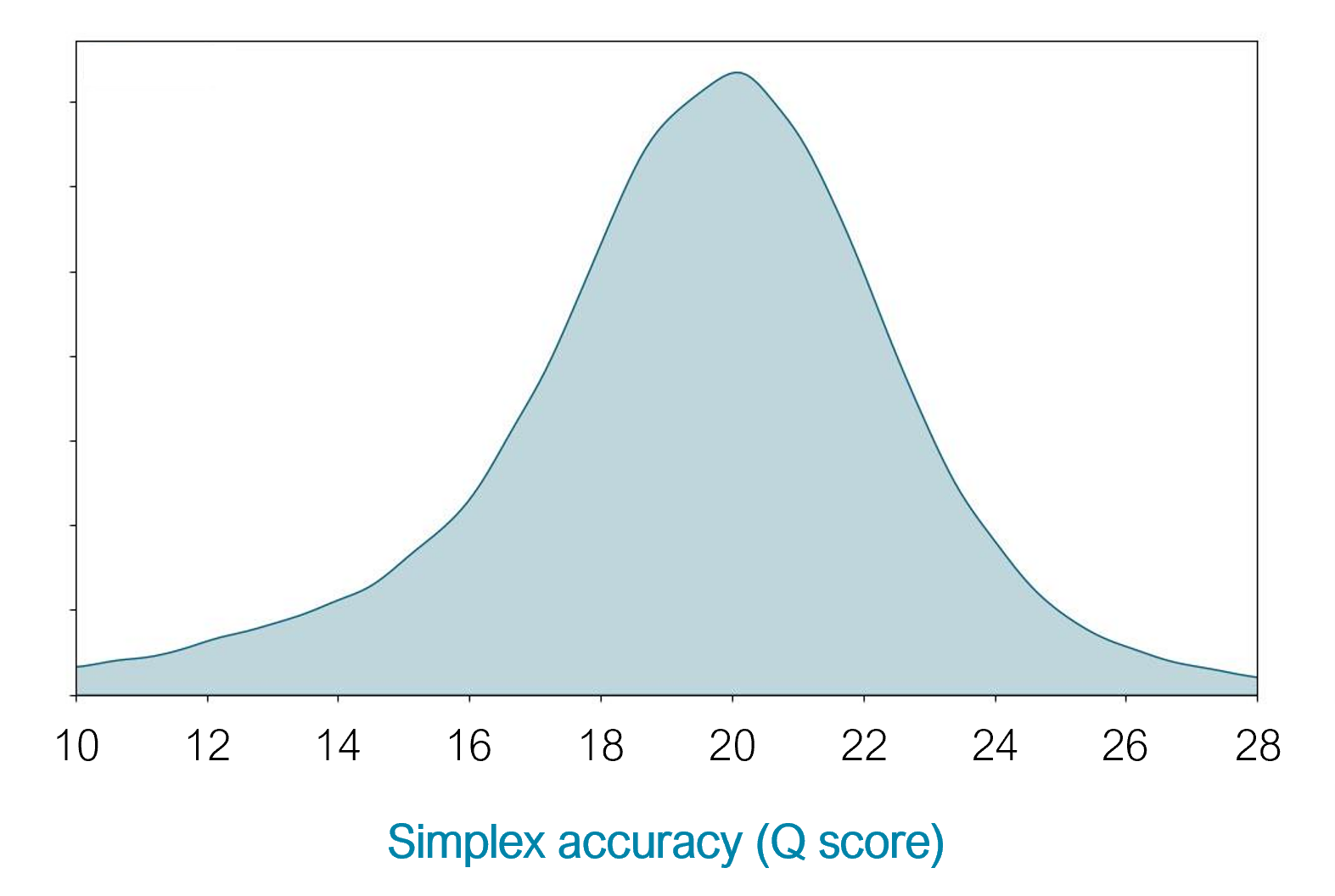
https://nanoporetech.com/q20plus-chemistry
10.4.1 flowcells + v14 chemistry: duplex
- The quality is even higher for duplex reads: mean of 30
- BUT: less than 50% of reads are duplex (i.e., you don’t always manage to read both strands)
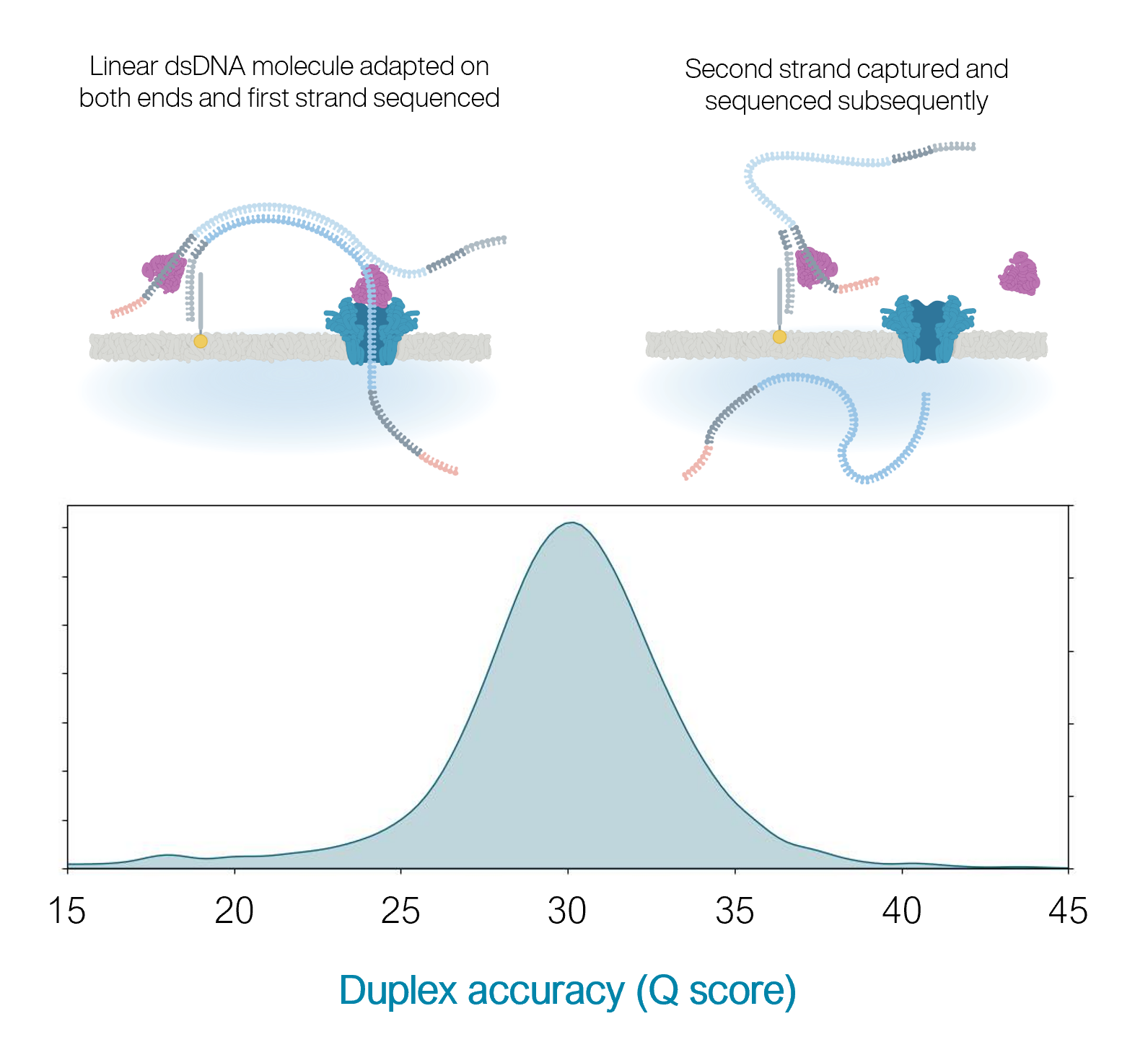
https://nanoporetech.com/q20plus-chemistry
More Nanopore: London Calling 2023
- Online conference held in May 2023
- Talk videos available online
- LOTS of really cool announcements and research applications
- Can also watch presentations from previous years.
Key Points
ONT produce a range of popular sequencing platforms.
This technology is constantly advancing.